Comparing individual-based approaches to modelling the self-organization of multicellular tissues
This section contains pages generated automatically from the source code accompanying the paper “Comparing individual-based approaches to modelling the self-organization of multicellular tissues”, PLOS Comput. Biol., 2017, doi: 10.1371/journal.pcbi.1005387.
Videos
Adhesion:
Proliferation
Short-range signalling
Long-range signalling
Getting the code and installing dependencies
Before running these examples you will need to install Chaste’s dependencies. The easiest way to do this is using an Ubuntu machine (or an Ubuntu virtual machine) as discussed on InstallGuides/UbuntuPackage. Note that Chaste is only fully supported on Linux/Unix systems, so users of Windows or Mac OS X are encouraged to follow the Docker instructions.
To checkout the source code for the specific code revision use the command
git clone https://github.com/Chaste/Chaste.git Chaste
cd ChasteThis project must be built with CMake. At this point you should configure Chaste with CMake.
You will also need the (updated-since-publication) source for the CellBasedComparison2017 project. This can be done by checking out the version from the repository by using the command
cd projects
git clone https://github.com/Chaste/CellBasedComparison2017.gitNow the project should be installed, and everything should compile and run correctly. You can now run the tests or simulations, or create your own test suites.
Documentation
There are two folders - src and test.
- The
srcfolder contains classes which add functionality to the core Chaste code. - The
testfolder contains:- TestCellSortingLiteratePaper.hpp - this file can be run to generate the results in Figures 2 and 3.
- TestCylindricalCryptLiteratePaper.hpp - this file can be run to generate the results in Figures 4, 5 and 6.
- TestDeltaNotchLiteratePaper.hpp - this file can be run to generate the results in Figures 7 and 8.
- TestMorphogenMonolayerLiteratePaper.hpp - this file can be run to generate the results in Figures 9, 10 and 11.
Clicking on the images below will take you to the code to run the corresponding simulation.
| Adhesion | Proliferation | Short-range signalling | Long-range signalling | |
|---|---|---|---|---|
| CA | 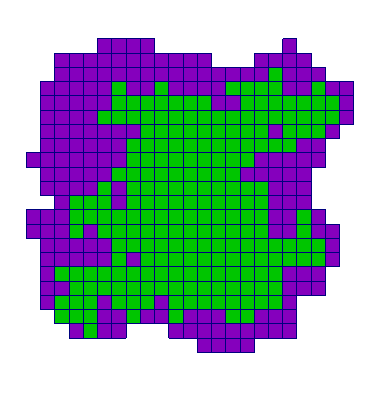 | 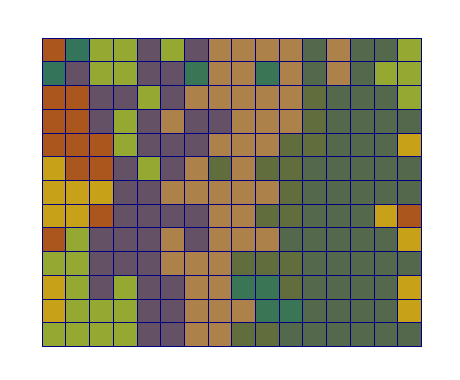 | 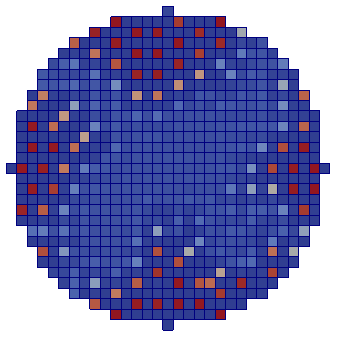 |  |
| CP | 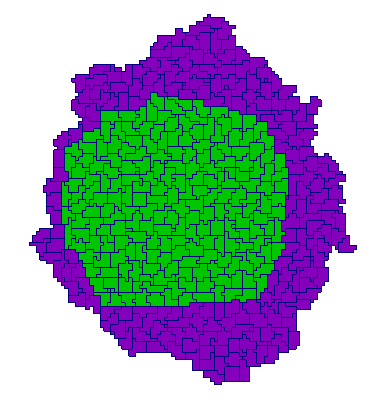 |  |  |  |
| OS |  |  |  |  |
| VT | 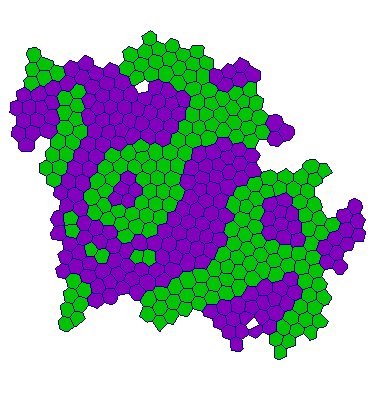 | 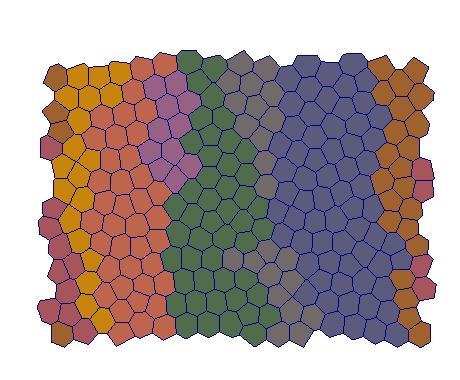 | 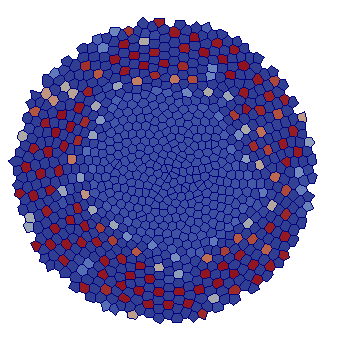 |  |
| VM | 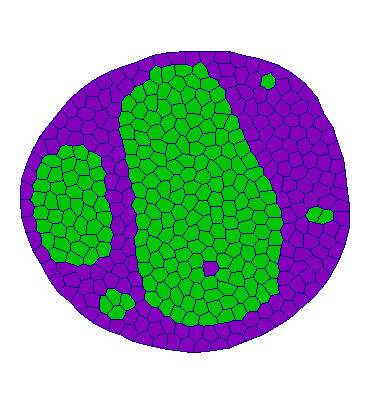 | 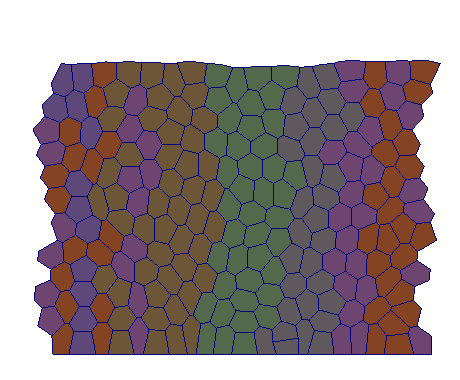 | 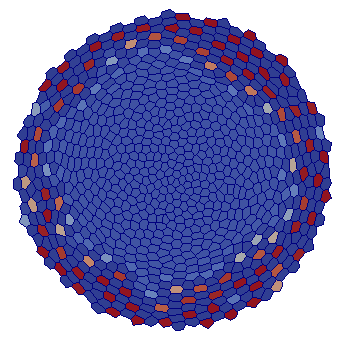 |  |
Running tests
To run the tests, starting from the main Chaste source directory:
mkdir chaste-build && cd chaste-build
cmake ..
## Make and run a test at a time:
make TestCellSortingLiteratePaperRunner
ctest -R TestCellSortingLiteratePaper
## etc.
## Make and run all tests on 4 threads:
make -j 4 project_CellBasedComparison2017
ctest -j 4 -L project_CellBasedComparison2017For further information on using Chaste, see the extensive guide material. You may also wish to look at some of the user tutorials.
