Cell-based simulation: tumour spheroid with oxygen diffusion and uptake
On this page
Note that the code is given in full at the bottom of the page.
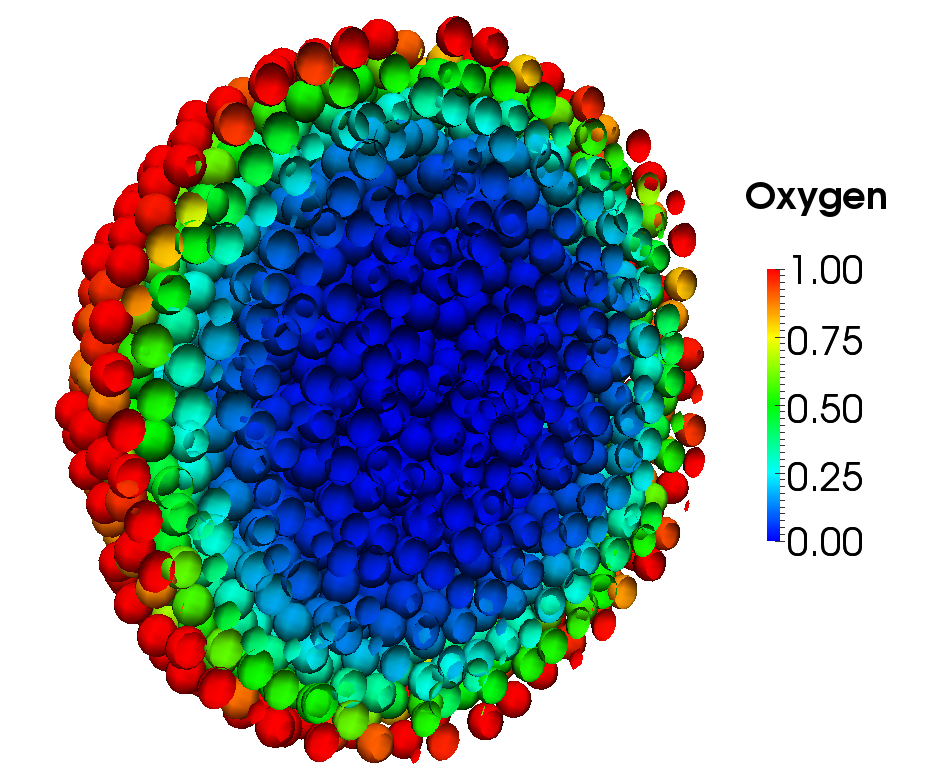
On this wiki page we describe in detail the code that is used to run this example from the paper.
The simulation is a mesh-based off-lattice simulation. In other words, cells are represented by points in space (nodes of a mesh) and are allowed to move without being confined to certain lattice sites.
We show a hybrid discrete-continuum model by using a reaction-diffusion PDE for oxygen, which discrete cells then respond to be proliferating in high oxygen, or dying in low oxygen.
We also show how a simulation can be checkpointed: that is, written to disk, reloaded, and continued.
This example uses only files from the core repository.
Remember to run with build=GccOptNative for speed.
e.g.
scons build=GccOptNative test_suite=projects/project_Plos2013/test/TestSpheroidExperimentsLiteratePaper.hppThe easiest way to visualize this simulation is with paraview.
Code overview
The first thing to do is to include the necessary header files.
#include <cxxtest/TestSuite.h>
// Must be included before other cell_based headers
#include "CellBasedSimulationArchiver.hpp"
#include <iomanip>
#include <boost/foreach.hpp>
#include "OffLatticeSimulation.hpp"
#include "CellBasedEventHandler.hpp"
#include "MeshBasedCellPopulation.hpp"
#include "NodeBasedCellPopulation.hpp"
#include "StochasticOxygenBasedCellCycleModel.hpp"
#include "GeneralisedLinearSpringForce.hpp"
#include "AveragedSourcePde.hpp"
#include "CellwiseSourcePde.hpp"
#include "ConstBoundaryCondition.hpp"
#include "CellBasedPdeHandler.hpp"
#include "ChastePoint.hpp"
#include "SmartPointers.hpp"
#include "ApoptoticCellKiller.hpp"
#include "AbstractCellBasedTestSuite.hpp"
#include "PetscSetupAndFinalize.hpp"
class TestSpheroidExperimentsLiteratePaper : public AbstractCellBasedTestSuite
{
private:These methods are cxx-test instructions running before and after each test below. They just report the time the test took, in different parts of the code.
void setUp()
{
AbstractCellBasedTestSuite::setUp();
CellBasedEventHandler::Reset();
}
void tearDown()
{
AbstractCellBasedTestSuite::tearDown();
CellBasedEventHandler::Headings();
CellBasedEventHandler::Report();
}
public:This example is split into two separate tests/simulations to demonstrate the check-pointing abilities of Chaste.
The first test runs from t=0 to t=100, and the second from t=100 to t=150.
It could equally well be reproduced by setting the end time in the first test to 150.
void TestMeshBasedSpheroidWithPde() throw(Exception)
{Create a simple 3D mesh, initially comprised of just five nodes
std::vector<Node<3>*> nodes;
nodes.push_back(new Node<3>(0, true, 0.0, 0.0, 0.0));
nodes.push_back(new Node<3>(1, true, 1.0, 1.0, 0.0));
nodes.push_back(new Node<3>(2, true, 1.0, 0.0, 1.0));
nodes.push_back(new Node<3>(3, true, 0.0, 1.0, 1.0));
nodes.push_back(new Node<3>(4, false, 0.5, 0.5, 0.5));
MutableMesh<3,3> mesh(nodes);Set up cells for each of the nodes in the mesh
boost::shared_ptr<AbstractCellMutationState> p_state(new WildTypeCellMutationState);
// We use the stem cell G1 duration, so make these 'stem' cells
MAKE_PTR(StemCellProliferativeType, p_stem_type);
std::vector<CellPtr> cells;
for (unsigned i=0; i<nodes.size(); i++)
{
StochasticOxygenBasedCellCycleModel* p_model = new StochasticOxygenBasedCellCycleModel();
p_model->SetDimension(3);We alter a number of parameters from their defaults, to speed up this example.
p_model->SetStemCellG1Duration(4.0);
p_model->SetHypoxicConcentration(0.1);
p_model->SetQuiescentConcentration(0.3);
p_model->SetCriticalHypoxicDuration(8);
double birth_time = -RandomNumberGenerator::Instance()->ranf()*
( p_model->GetStemCellG1Duration()
+ p_model->GetSG2MDuration() );
CellPtr p_cell(new Cell(p_state, p_model));
p_cell->SetBirthTime(birth_time);
p_cell->SetCellProliferativeType(p_stem_type);
cells.push_back(p_cell);
}Create cell population - a mapping between a mesh and cells.
MeshBasedCellPopulation<3> cell_population(mesh, cells);
cell_population.SetAbsoluteMovementThreshold(DBL_MAX);
cell_population.SetWriteVtkAsPoints(true);Set up cell data on the cell population
cell_population.SetDataOnAllCells("oxygen", 1.0);Set up cell-based simulation
OffLatticeSimulation<3> simulator(cell_population);
simulator.SetEndTime(100); // hours
Default time step is 30 seconds, so this gives two visualisation outputs each hour.
simulator.SetSamplingTimestepMultiple(60);
simulator.SetOutputDirectory("Plos2013_MeshBasedSpheroidWithPde");Set up PDE and boundary conditions
CellwiseSourcePde<3> pde(cell_population, -1.0);
ConstBoundaryCondition<3> bc(1.0);
bool is_neumann_bc = false;
PdeAndBoundaryConditions<3> pde_and_bc(&pde, &bc, is_neumann_bc);
pde_and_bc.SetDependentVariableName("oxygen");Create a handler (for any number of PDEs+BCs, in this case we just add one).
CellBasedPdeHandler<3> pde_handler(&cell_population);
pde_handler.AddPdeAndBc(&pde_and_bc);Pass PDE handler to the simulation
simulator.SetCellBasedPdeHandler(&pde_handler);Create a force law and pass it to the simulation
MAKE_PTR(GeneralisedLinearSpringForce<3>, p_force);
p_force->SetMeinekeSpringStiffness(30.0); // default is 15.0;
p_force->SetCutOffLength(1.5);
simulator.AddForce(p_force);Set up cell killer and pass into simulation.
In this simulation the cell cycle model gives cells an
ApoptoticCellProperty if they are in low oxygen (as
defined by the PDE). This cell killer removes cells that
have this property.
MAKE_PTR_ARGS(ApoptoticCellKiller<3>, p_killer, (&cell_population));
simulator.AddCellKiller(p_killer);Run the simulation
simulator.Solve();Save the results
CellBasedSimulationArchiver<3, OffLatticeSimulation<3> >::Save(&simulator);
}
void TestLongerMeshBasedSpheroidWithPde() throw(Exception)
{The archive is to be copied from previous test output It could be stored and re-loaded from anywhere you like.
This is useful for checkpointing on large HPC machines, and also if you want to experiment with different interventions on an existing spheroid state.
FileFinder test_data_directory("Plos2013_MeshBasedSpheroidWithPde/archive",
RelativeTo::ChasteTestOutput);We specify a location to copy the archive files from the previous test to
OutputFileHandler archive_handler("Plos2013_LongerMeshBasedSpheroidWithPde/archive");And copy the files across (this uses the boost filesystem library, incidentally)
// Following is done in two lines to avoid a bug in Intel compiler v12.0!
std::vector<FileFinder> temp_files = test_data_directory.FindMatches("*");
BOOST_FOREACH(FileFinder temp_file, temp_files)
{
archive_handler.CopyFileTo(temp_file);
}Load the simulation up from 100 hours archive
OffLatticeSimulation<3>* p_simulator
= CellBasedSimulationArchiver<3, OffLatticeSimulation<3> >::Load("Plos2013_LongerMeshBasedSpheroidWithPde", 100);Change some settings, a new end time and output directory
p_simulator->SetEndTime(150);
p_simulator->SetOutputDirectory("Plos2013_LongerMeshBasedSpheroidWithPde");Run the simulation to the new end time
p_simulator->Solve();Save the results
CellBasedSimulationArchiver<3, OffLatticeSimulation<3> >::Save(p_simulator);
}
};Full code
#include <cxxtest/TestSuite.h>
// Must be included before other cell_based headers
#include "CellBasedSimulationArchiver.hpp"
#include <iomanip>
#include <boost/foreach.hpp>
#include "OffLatticeSimulation.hpp"
#include "CellBasedEventHandler.hpp"
#include "MeshBasedCellPopulation.hpp"
#include "NodeBasedCellPopulation.hpp"
#include "StochasticOxygenBasedCellCycleModel.hpp"
#include "GeneralisedLinearSpringForce.hpp"
#include "AveragedSourcePde.hpp"
#include "CellwiseSourcePde.hpp"
#include "ConstBoundaryCondition.hpp"
#include "CellBasedPdeHandler.hpp"
#include "ChastePoint.hpp"
#include "SmartPointers.hpp"
#include "ApoptoticCellKiller.hpp"
#include "AbstractCellBasedTestSuite.hpp"
#include "PetscSetupAndFinalize.hpp"
class TestSpheroidExperimentsLiteratePaper : public AbstractCellBasedTestSuite
{
private:
void setUp()
{
AbstractCellBasedTestSuite::setUp();
CellBasedEventHandler::Reset();
}
void tearDown()
{
AbstractCellBasedTestSuite::tearDown();
CellBasedEventHandler::Headings();
CellBasedEventHandler::Report();
}
public:
void TestMeshBasedSpheroidWithPde() throw(Exception)
{
std::vector<Node<3>*> nodes;
nodes.push_back(new Node<3>(0, true, 0.0, 0.0, 0.0));
nodes.push_back(new Node<3>(1, true, 1.0, 1.0, 0.0));
nodes.push_back(new Node<3>(2, true, 1.0, 0.0, 1.0));
nodes.push_back(new Node<3>(3, true, 0.0, 1.0, 1.0));
nodes.push_back(new Node<3>(4, false, 0.5, 0.5, 0.5));
MutableMesh<3,3> mesh(nodes);
boost::shared_ptr<AbstractCellMutationState> p_state(new WildTypeCellMutationState);
// We use the stem cell G1 duration, so make these 'stem' cells
MAKE_PTR(StemCellProliferativeType, p_stem_type);
std::vector<CellPtr> cells;
for (unsigned i=0; i<nodes.size(); i++)
{
StochasticOxygenBasedCellCycleModel* p_model = new StochasticOxygenBasedCellCycleModel();
p_model->SetDimension(3);
p_model->SetStemCellG1Duration(4.0);
p_model->SetHypoxicConcentration(0.1);
p_model->SetQuiescentConcentration(0.3);
p_model->SetCriticalHypoxicDuration(8);
double birth_time = -RandomNumberGenerator::Instance()->ranf()*
( p_model->GetStemCellG1Duration()
+ p_model->GetSG2MDuration() );
CellPtr p_cell(new Cell(p_state, p_model));
p_cell->SetBirthTime(birth_time);
p_cell->SetCellProliferativeType(p_stem_type);
cells.push_back(p_cell);
}
MeshBasedCellPopulation<3> cell_population(mesh, cells);
cell_population.SetAbsoluteMovementThreshold(DBL_MAX);
cell_population.SetWriteVtkAsPoints(true);
cell_population.SetDataOnAllCells("oxygen", 1.0);
OffLatticeSimulation<3> simulator(cell_population);
simulator.SetEndTime(100); // hours
simulator.SetSamplingTimestepMultiple(60);
simulator.SetOutputDirectory("Plos2013_MeshBasedSpheroidWithPde");
CellwiseSourcePde<3> pde(cell_population, -1.0);
ConstBoundaryCondition<3> bc(1.0);
bool is_neumann_bc = false;
PdeAndBoundaryConditions<3> pde_and_bc(&pde, &bc, is_neumann_bc);
pde_and_bc.SetDependentVariableName("oxygen");
CellBasedPdeHandler<3> pde_handler(&cell_population);
pde_handler.AddPdeAndBc(&pde_and_bc);
simulator.SetCellBasedPdeHandler(&pde_handler);
MAKE_PTR(GeneralisedLinearSpringForce<3>, p_force);
p_force->SetMeinekeSpringStiffness(30.0); // default is 15.0;
p_force->SetCutOffLength(1.5);
simulator.AddForce(p_force);
MAKE_PTR_ARGS(ApoptoticCellKiller<3>, p_killer, (&cell_population));
simulator.AddCellKiller(p_killer);
simulator.Solve();
CellBasedSimulationArchiver<3, OffLatticeSimulation<3> >::Save(&simulator);
}
void TestLongerMeshBasedSpheroidWithPde() throw(Exception)
{
FileFinder test_data_directory("Plos2013_MeshBasedSpheroidWithPde/archive",
RelativeTo::ChasteTestOutput);
OutputFileHandler archive_handler("Plos2013_LongerMeshBasedSpheroidWithPde/archive");
// Following is done in two lines to avoid a bug in Intel compiler v12.0!
std::vector<FileFinder> temp_files = test_data_directory.FindMatches("*");
BOOST_FOREACH(FileFinder temp_file, temp_files)
{
archive_handler.CopyFileTo(temp_file);
}
OffLatticeSimulation<3>* p_simulator
= CellBasedSimulationArchiver<3, OffLatticeSimulation<3> >::Load("Plos2013_LongerMeshBasedSpheroidWithPde", 100);
p_simulator->SetEndTime(150);
p_simulator->SetOutputDirectory("Plos2013_LongerMeshBasedSpheroidWithPde");
p_simulator->Solve();
CellBasedSimulationArchiver<3, OffLatticeSimulation<3> >::Save(p_simulator);
}
};