Documentation for Release 2024.2
Lattice Based Angiogenesis
This tutorial is designed to introduce a lattice based angiogenesis problem based on a simplified version of the vascular tumour application described in Owen et al. 2011. It is a 2D simulation using cellular automaton for cells, a regular grid for vessel movement and the same grid for the solution of partial differential equations for oxygen and VEGF transport using the finite difference method.
The Test
Start by introducing the necessary header files. The first contain functionality for setting up unit tests, smart pointer tools and output management,
#include <vector>
#include "SmartPointers.hpp"
#include "OutputFileHandler.hpp"
#include "AbstractCellBasedWithTimingsTestSuite.hpp"
#include "RandomNumberGenerator.hpp"dimensional analysis,
#include "DimensionalChastePoint.hpp"
#include "UnitCollection.hpp"
#include "Owen11Parameters.hpp"
#include "Secomb04Parameters.hpp"
#include "GenericParameters.hpp"
#include "ParameterCollection.hpp"
#include "BaseUnits.hpp"vessel networks,
#include "VesselNode.hpp"
#include "VesselNetwork.hpp"cells,
#include "CancerCellMutationState.hpp"
#include "StalkCellMutationState.hpp"
#include "QuiescentCancerCellMutationState.hpp"
#include "WildTypeCellMutationState.hpp"
#include "Owen11CellPopulationGenerator.hpp"
#include "Owen2011TrackingModifier.hpp"
#include "CaBasedCellPopulation.hpp"
#include "ApoptoticCellKiller.hpp"flow,
#include "VesselImpedanceCalculator.hpp"
#include "FlowSolver.hpp"
#include "ConstantHaematocritSolver.hpp"
#include "StructuralAdaptationSolver.hpp"
#include "WallShearStressCalculator.hpp"
#include "MechanicalStimulusCalculator.hpp"
#include "MetabolicStimulusCalculator.hpp"
#include "ShrinkingStimulusCalculator.hpp"
#include "ViscosityCalculator.hpp"grids and PDEs,
#include "RegularGrid.hpp"
#include "FiniteDifferenceSolver.hpp"
#include "CellBasedDiscreteSource.hpp"
#include "VesselBasedDiscreteSource.hpp"
#include "CellStateDependentDiscreteSource.hpp"
#include "DiscreteContinuumBoundaryCondition.hpp"
#include "LinearSteadyStateDiffusionReactionPde.hpp"angiogenesis and regression,
#include "Owen2011SproutingRule.hpp"
#include "Owen2011MigrationRule.hpp"
#include "AngiogenesisSolver.hpp"
#include "WallShearStressBasedRegressionSolver.hpp"and classes for managing the simulation.
#include "MicrovesselSolver.hpp"
#include "MicrovesselSimulationModifier.hpp"
#include "OnLatticeSimulation.hpp"This should appear last.
#include "PetscSetupAndFinalize.hpp"
class TestLatticeBasedAngiogenesisLiteratePaper : public AbstractCellBasedWithTimingsTestSuite
{
public:
void Test2dLatticeBased() throw (Exception)
{Set up output file management and seed the random number generator.
MAKE_PTR_ARGS(OutputFileHandler, p_handler, ("TestLatticeBasedAngiogenesisLiteratePaper"));
RandomNumberGenerator::Instance()->Reseed(12345);This component uses explicit dimensions for all quantities, but interfaces with solvers which take
non-dimensional inputs. The BaseUnits singleton takes time, length and mass reference scales to
allow non-dimensionalisation when sending quantities to external solvers and re-dimensionalisation of
results. For our purposes microns for length and hours for time are suitable base units.
units::quantity<unit::length> reference_length(1.0 * unit::microns);
units::quantity<unit::time> reference_time(1.0* unit::hours);
BaseUnits::Instance()->SetReferenceLengthScale(reference_length);
BaseUnits::Instance()->SetReferenceTimeScale(reference_time);Set up the lattice (grid), we will use the same dimensions as Owen et al. 2011.
Note that we are using hard-coded parameters from that paper. You can see the values by inspecting Owen11Parameters.cpp.
Alternatively each parameter supports the << operator for streaming. When we get the value of the parameter by doing
Owen11Parameters::mpLatticeSpacing->GetValue("User") a record is kept that this parameter has been used in the simulation.
A record of all parameters used in a simulation can be dumped to file on completion, as will be shown below.
boost::shared_ptr<RegularGrid<2> > p_grid = RegularGrid<2>::Create();
units::quantity<unit::length> grid_spacing = Owen11Parameters::mpLatticeSpacing->GetValue("User");
p_grid->SetSpacing(grid_spacing);
std::vector<unsigned> extents(3, 1);
extents[0] = 51; // num x
extents[1] = 51; // num_y
p_grid->SetExtents(extents);We can write the lattice to file for quick visualization with Paraview. Rendering of this and subsequent images is performed using standard Paraview operations, not detailed here.
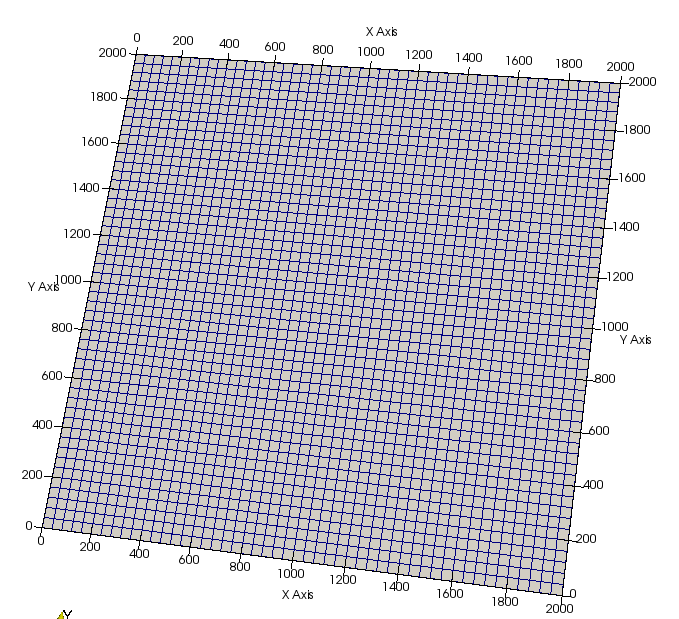
p_grid->Write(p_handler);Next, set up the vessel network, this will initially consist of two, large counter-flowing vessels. Also set the inlet and outlet pressures and flags.
boost::shared_ptr<VesselNode<2> > p_node11 = VesselNode<2>::Create(0.0, 400.0, 0.0, reference_length);
boost::shared_ptr<VesselNode<2> > p_node12 = VesselNode<2>::Create(2000.0, 400.0, 0.0, reference_length);
p_node11->GetFlowProperties()->SetIsInputNode(true);
p_node11->GetFlowProperties()->SetPressure(Owen11Parameters::mpInletPressure->GetValue("User"));
p_node12->GetFlowProperties()->SetIsOutputNode(true);
p_node12->GetFlowProperties()->SetPressure(Owen11Parameters::mpOutletPressure->GetValue("User"));
boost::shared_ptr<VesselNode<2> > p_node13 = VesselNode<2>::Create(2000.0, 1600.0, 0.0, reference_length);
boost::shared_ptr<VesselNode<2> > p_node14 = VesselNode<2>::Create(0.0, 1600.0, 0.0, reference_length);
p_node13->GetFlowProperties()->SetIsInputNode(true);
p_node13->GetFlowProperties()->SetPressure(Owen11Parameters::mpInletPressure->GetValue("User"));
p_node14->GetFlowProperties()->SetIsOutputNode(true);
p_node14->GetFlowProperties()->SetPressure(Owen11Parameters::mpOutletPressure->GetValue("User"));
boost::shared_ptr<Vessel<2> > p_vessel1 = Vessel<2>::Create(p_node11, p_node12);
boost::shared_ptr<Vessel<2> > p_vessel2 = Vessel<2>::Create(p_node13, p_node14);
boost::shared_ptr<VesselNetwork<2> > p_network = VesselNetwork<2>::Create();
p_network->AddVessel(p_vessel1);
p_network->AddVessel(p_vessel2);Again, we can write the network to file for quick visualization with Paraview.
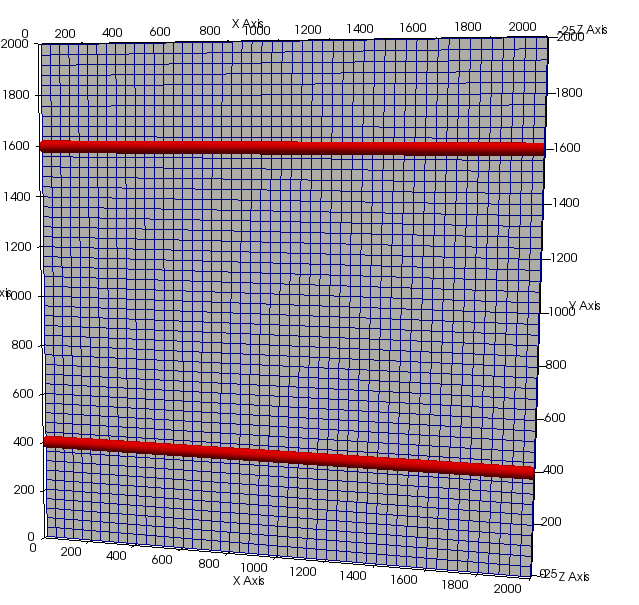
p_network->Write(p_handler->GetOutputDirectoryFullPath() + "initial_network.vtp");Next, set up the cell populations. We will setup up a population similar to that used in the Owen et al., 2011 paper. That is, a grid filled with normal cells and a tumour spheroid in the middle. We can use a generator for this purpose. The generator simply sets up the population using conventional Cell Based Chaste methods.
boost::shared_ptr<Owen11CellPopulationGenerator<2> > p_cell_population_genenerator = Owen11CellPopulationGenerator<2>::Create();
p_cell_population_genenerator->SetRegularGrid(p_grid);
p_cell_population_genenerator->SetVesselNetwork(p_network);
units::quantity<unit::length> tumour_radius(300.0 * unit::microns);
p_cell_population_genenerator->SetTumourRadius(tumour_radius);
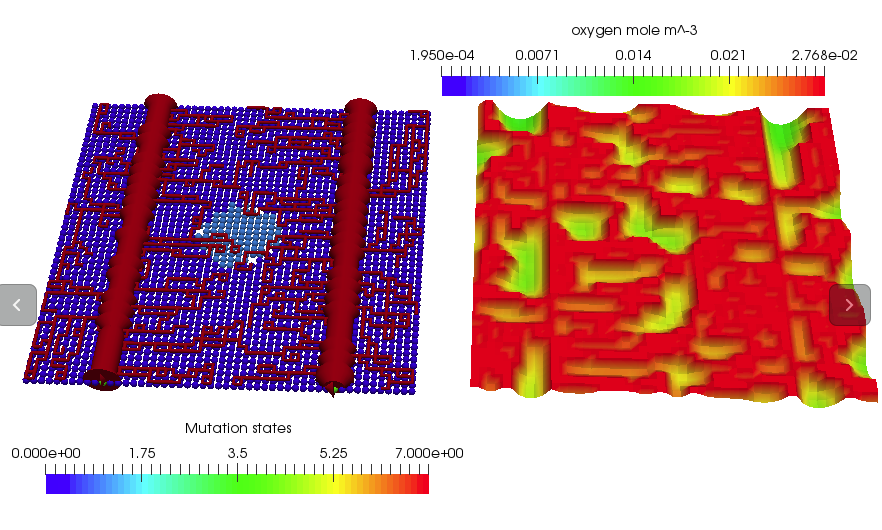
boost::shared_ptr<CaBasedCellPopulation<2> > p_cell_population = p_cell_population_genenerator->Update();At this point the simulation domain will look as follows:
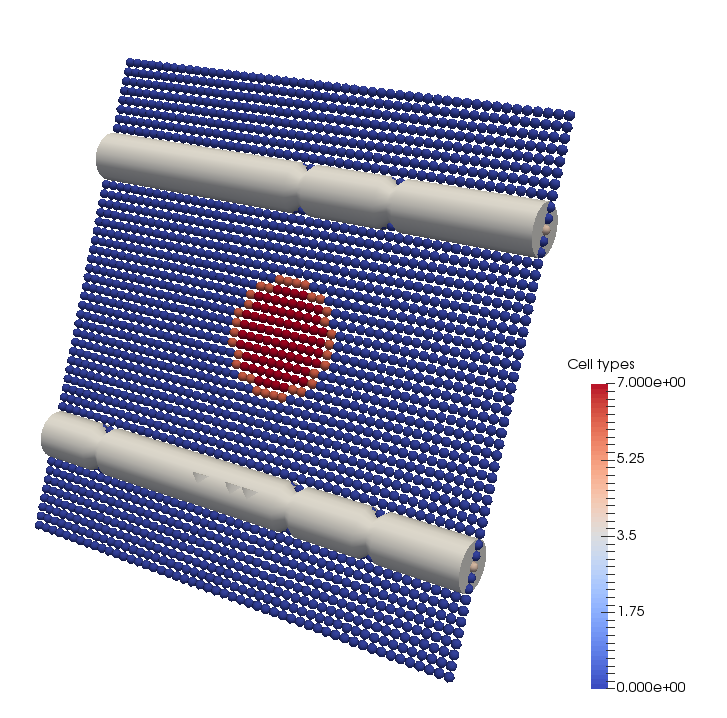
Next set up the PDEs for oxygen and VEGF. Cells will act as discrete oxygen sinks and discrete vegf sources. A sample PDE solution for oxygen is shown below:
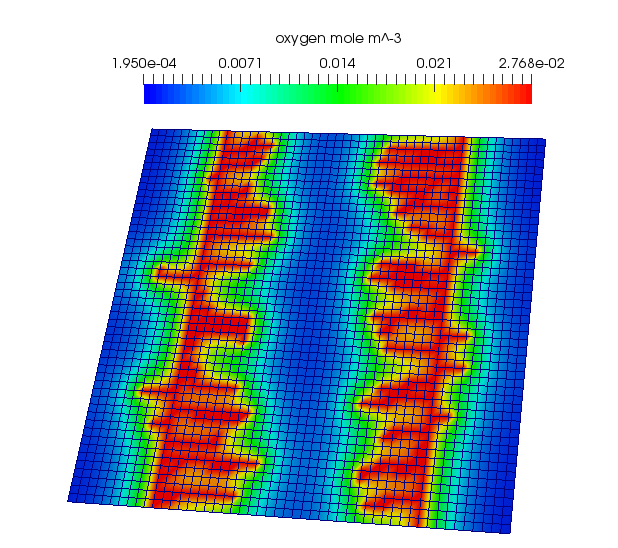
boost::shared_ptr<LinearSteadyStateDiffusionReactionPde<2> > p_oxygen_pde = LinearSteadyStateDiffusionReactionPde<2>::Create();
p_oxygen_pde->SetIsotropicDiffusionConstant(Owen11Parameters::mpOxygenDiffusivity->GetValue("User"));
boost::shared_ptr<CellBasedDiscreteSource<2> > p_cell_oxygen_sink = CellBasedDiscreteSource<2>::Create();
p_cell_oxygen_sink->SetLinearInUConsumptionRatePerCell(Owen11Parameters::mpCellOxygenConsumptionRate->GetValue("User"));
p_oxygen_pde->AddDiscreteSource(p_cell_oxygen_sink);Vessels release oxygen depending on their haematocrit levels
boost::shared_ptr<VesselBasedDiscreteSource<2> > p_vessel_oxygen_source = VesselBasedDiscreteSource<2>::Create();
units::quantity<unit::solubility> oxygen_solubility_at_stp = Secomb04Parameters::mpOxygenVolumetricSolubility->GetValue("User") *
GenericParameters::mpGasConcentrationAtStp->GetValue("User");
units::quantity<unit::concentration> vessel_oxygen_concentration = oxygen_solubility_at_stp *
Owen11Parameters::mpReferencePartialPressure->GetValue("User");
p_vessel_oxygen_source->SetReferenceConcentration(vessel_oxygen_concentration);
p_vessel_oxygen_source->SetVesselPermeability(Owen11Parameters::mpVesselOxygenPermeability->GetValue("User"));
p_vessel_oxygen_source->SetReferenceHaematocrit(Owen11Parameters::mpInflowHaematocrit->GetValue("User"));
p_oxygen_pde->AddDiscreteSource(p_vessel_oxygen_source);Set up a finite difference solver and pass it the pde and grid.
boost::shared_ptr<FiniteDifferenceSolver<2> > p_oxygen_solver = FiniteDifferenceSolver<2>::Create();
p_oxygen_solver->SetPde(p_oxygen_pde);
p_oxygen_solver->SetLabel("oxygen");
p_oxygen_solver->SetGrid(p_grid);The rate of VEGF release depends on the cell type and intracellular VEGF levels, so we need a more detailed type of discrete source. A sample PDE solution for VEGF is shown below.
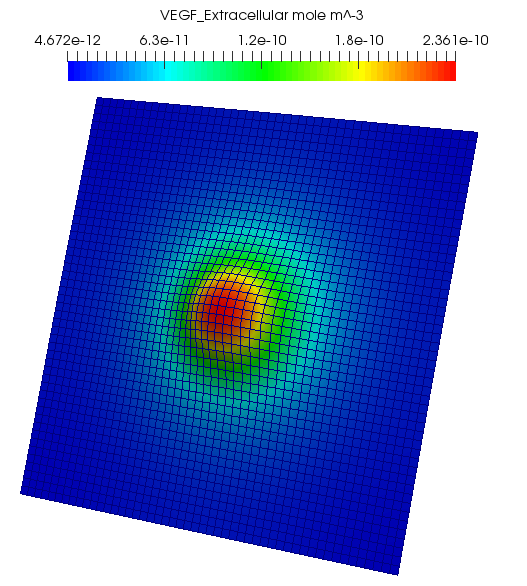
boost::shared_ptr<LinearSteadyStateDiffusionReactionPde<2> > p_vegf_pde = LinearSteadyStateDiffusionReactionPde<2>::Create();
p_vegf_pde->SetIsotropicDiffusionConstant(Owen11Parameters::mpVegfDiffusivity->GetValue("User"));
p_vegf_pde->SetContinuumLinearInUTerm(-Owen11Parameters::mpVegfDecayRate->GetValue("User"));Set up a map for different release rates depending on cell type. Also include a threshold intracellular VEGF below which there is no release.
boost::shared_ptr<CellStateDependentDiscreteSource<2> > p_normal_and_quiescent_cell_source = CellStateDependentDiscreteSource<2>::Create();
std::map<unsigned, units::quantity<unit::concentration_flow_rate> > normal_and_quiescent_cell_rates;
std::map<unsigned, units::quantity<unit::concentration> > normal_and_quiescent_cell_rate_thresholds;
MAKE_PTR(QuiescentCancerCellMutationState, p_quiescent_cancer_state);
MAKE_PTR(WildTypeCellMutationState, p_normal_cell_state);
normal_and_quiescent_cell_rates[p_normal_cell_state->GetColour()] = Owen11Parameters::mpCellVegfSecretionRate->GetValue("User");
normal_and_quiescent_cell_rate_thresholds[p_normal_cell_state->GetColour()] = 0.27*unit::mole_per_metre_cubed;
normal_and_quiescent_cell_rates[p_quiescent_cancer_state->GetColour()] = Owen11Parameters::mpCellVegfSecretionRate->GetValue("User");
normal_and_quiescent_cell_rate_thresholds[p_quiescent_cancer_state->GetColour()] = 0.0*unit::mole_per_metre_cubed;
p_normal_and_quiescent_cell_source->SetStateRateMap(normal_and_quiescent_cell_rates);
p_normal_and_quiescent_cell_source->SetLabelName("VEGF");
p_normal_and_quiescent_cell_source->SetStateRateThresholdMap(normal_and_quiescent_cell_rate_thresholds);
p_vegf_pde->AddDiscreteSource(p_normal_and_quiescent_cell_source);Add a vessel related VEGF sink
boost::shared_ptr<VesselBasedDiscreteSource<2> > p_vessel_vegf_sink = VesselBasedDiscreteSource<2>::Create();
p_vessel_vegf_sink->SetReferenceConcentration(0.0*unit::mole_per_metre_cubed);
p_vessel_vegf_sink->SetVesselPermeability(Owen11Parameters::mpVesselVegfPermeability->GetValue("User"));
p_vegf_pde->AddDiscreteSource(p_vessel_vegf_sink);Set up a finite difference solver as before.
boost::shared_ptr<FiniteDifferenceSolver<2> > p_vegf_solver = FiniteDifferenceSolver<2>::Create();
p_vegf_solver->SetPde(p_vegf_pde);
p_vegf_solver->SetLabel("VEGF_Extracellular");
p_vegf_solver->SetGrid(p_grid);Next set up the flow problem. Assign a blood plasma viscosity to the vessels. The actual viscosity will depend on haematocrit and diameter. This solver manages growth and shrinkage of vessels in response to flow related stimuli. A sample plot of the stimulus distrbution during a simulation is shown below:

units::quantity<unit::length> large_vessel_radius(25.0 * unit::microns);
p_network->SetSegmentRadii(large_vessel_radius);
units::quantity<unit::dynamic_viscosity> viscosity = Owen11Parameters::mpPlasmaViscosity->GetValue("User");
p_network->SetSegmentViscosity(viscosity);Set up the pre- and post flow calculators.
boost::shared_ptr<VesselImpedanceCalculator<2> > p_impedance_calculator = VesselImpedanceCalculator<2>::Create();
boost::shared_ptr<ConstantHaematocritSolver<2> > p_haematocrit_calculator = ConstantHaematocritSolver<2>::Create();
p_haematocrit_calculator->SetHaematocrit(Owen11Parameters::mpInflowHaematocrit->GetValue("User"));
boost::shared_ptr<WallShearStressCalculator<2> > p_wss_calculator = WallShearStressCalculator<2>::Create();
boost::shared_ptr<MechanicalStimulusCalculator<2> > p_mech_stimulus_calculator = MechanicalStimulusCalculator<2>::Create();
boost::shared_ptr<MetabolicStimulusCalculator<2> > p_metabolic_stim_calculator = MetabolicStimulusCalculator<2>::Create();
boost::shared_ptr<ShrinkingStimulusCalculator<2> > p_shrinking_stimulus_calculator = ShrinkingStimulusCalculator<2>::Create();
boost::shared_ptr<ViscosityCalculator<2> > p_viscosity_calculator = ViscosityCalculator<2>::Create();Set up and configure the structural adaptation solver.
boost::shared_ptr<StructuralAdaptationSolver<2> > p_structural_adaptation_solver = StructuralAdaptationSolver<2>::Create();
p_structural_adaptation_solver->SetTolerance(0.0001);
p_structural_adaptation_solver->SetMaxIterations(100);
p_structural_adaptation_solver->SetTimeIncrement(Owen11Parameters::mpVesselRadiusUpdateTimestep->GetValue("User"));
p_structural_adaptation_solver->AddPreFlowSolveCalculator(p_impedance_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_haematocrit_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_wss_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_metabolic_stim_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_mech_stimulus_calculator);
// p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_shrinking_stimulus_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_viscosity_calculator);Set up a regression solver.
boost::shared_ptr<WallShearStressBasedRegressionSolver<2> > p_regression_solver =
WallShearStressBasedRegressionSolver<2>::Create();Set up an angiogenesis solver and add sprouting and migration rules.
boost::shared_ptr<AngiogenesisSolver<2> > p_angiogenesis_solver = AngiogenesisSolver<2>::Create();
boost::shared_ptr<Owen2011SproutingRule<2> > p_sprouting_rule = Owen2011SproutingRule<2>::Create();
boost::shared_ptr<Owen2011MigrationRule<2> > p_migration_rule = Owen2011MigrationRule<2>::Create();
p_angiogenesis_solver->SetMigrationRule(p_migration_rule);
p_angiogenesis_solver->SetSproutingRule(p_sprouting_rule);
p_sprouting_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_migration_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_angiogenesis_solver->SetVesselGrid(p_grid);
p_angiogenesis_solver->SetVesselNetwork(p_network);The microvessel solver will manage all aspects of the vessel solve.
boost::shared_ptr<MicrovesselSolver<2> > p_microvessel_solver = MicrovesselSolver<2>::Create();
p_microvessel_solver->SetVesselNetwork(p_network);
p_microvessel_solver->SetOutputFrequency(5);
p_microvessel_solver->AddDiscreteContinuumSolver(p_oxygen_solver);
p_microvessel_solver->AddDiscreteContinuumSolver(p_vegf_solver);
p_microvessel_solver->SetStructuralAdaptationSolver(p_structural_adaptation_solver);
p_microvessel_solver->SetRegressionSolver(p_regression_solver);
p_microvessel_solver->SetAngiogenesisSolver(p_angiogenesis_solver);The microvessel solution modifier will link the vessel and cell solvers. We need to explicitly tell is which extracellular fields to update based on PDE solutions.
boost::shared_ptr<MicrovesselSimulationModifier<2> > p_microvessel_modifier = MicrovesselSimulationModifier<2>::Create();
p_microvessel_modifier->SetMicrovesselSolver(p_microvessel_solver);
std::vector<std::string> update_labels;
update_labels.push_back("oxygen");
update_labels.push_back("VEGF_Extracellular");
p_microvessel_modifier->SetCellDataUpdateLabels(update_labels);The full simulation is run as a typical Cell Based Chaste simulation
OnLatticeSimulation<2> simulator(*p_cell_population);
simulator.AddSimulationModifier(p_microvessel_modifier);Add a killer to remove apoptotic cells
boost::shared_ptr<ApoptoticCellKiller<2> > p_apoptotic_cell_killer(new ApoptoticCellKiller<2>(p_cell_population.get()));
simulator.AddCellKiller(p_apoptotic_cell_killer);Add another modifier for updating cell cycle quantities.
boost::shared_ptr<Owen2011TrackingModifier<2> > p_owen11_tracking_modifier(new Owen2011TrackingModifier<2>);
simulator.AddSimulationModifier(p_owen11_tracking_modifier);Set up the remainder of the simulation
simulator.SetOutputDirectory("TestLatticeBasedAngiogenesisLiteratePaper");
simulator.SetSamplingTimestepMultiple(5);
simulator.SetDt(0.5);This end time corresponds to roughly 10 minutes run-time on a desktop PC. Increase it or decrease as preferred. The end time used in Owen et al. 2011 is 4800 hours.
simulator.SetEndTime(20.0);Do the solve. A sample solution is shown at the top of this test.
simulator.Solve();Dump the parameters to file for inspection.
ParameterCollection::Instance()->DumpToFile(p_handler->GetOutputDirectoryFullPath()+"parameter_collection.xml");
}
};Full code
#include <vector>
#include "SmartPointers.hpp"
#include "OutputFileHandler.hpp"
#include "AbstractCellBasedWithTimingsTestSuite.hpp"
#include "RandomNumberGenerator.hpp"
#include "DimensionalChastePoint.hpp"
#include "UnitCollection.hpp"
#include "Owen11Parameters.hpp"
#include "Secomb04Parameters.hpp"
#include "GenericParameters.hpp"
#include "ParameterCollection.hpp"
#include "BaseUnits.hpp"
#include "VesselNode.hpp"
#include "VesselNetwork.hpp"
#include "CancerCellMutationState.hpp"
#include "StalkCellMutationState.hpp"
#include "QuiescentCancerCellMutationState.hpp"
#include "WildTypeCellMutationState.hpp"
#include "Owen11CellPopulationGenerator.hpp"
#include "Owen2011TrackingModifier.hpp"
#include "CaBasedCellPopulation.hpp"
#include "ApoptoticCellKiller.hpp"
#include "VesselImpedanceCalculator.hpp"
#include "FlowSolver.hpp"
#include "ConstantHaematocritSolver.hpp"
#include "StructuralAdaptationSolver.hpp"
#include "WallShearStressCalculator.hpp"
#include "MechanicalStimulusCalculator.hpp"
#include "MetabolicStimulusCalculator.hpp"
#include "ShrinkingStimulusCalculator.hpp"
#include "ViscosityCalculator.hpp"
#include "RegularGrid.hpp"
#include "FiniteDifferenceSolver.hpp"
#include "CellBasedDiscreteSource.hpp"
#include "VesselBasedDiscreteSource.hpp"
#include "CellStateDependentDiscreteSource.hpp"
#include "DiscreteContinuumBoundaryCondition.hpp"
#include "LinearSteadyStateDiffusionReactionPde.hpp"
#include "Owen2011SproutingRule.hpp"
#include "Owen2011MigrationRule.hpp"
#include "AngiogenesisSolver.hpp"
#include "WallShearStressBasedRegressionSolver.hpp"
#include "MicrovesselSolver.hpp"
#include "MicrovesselSimulationModifier.hpp"
#include "OnLatticeSimulation.hpp"
#include "PetscSetupAndFinalize.hpp"
class TestLatticeBasedAngiogenesisLiteratePaper : public AbstractCellBasedWithTimingsTestSuite
{
public:
void Test2dLatticeBased() throw (Exception)
{
MAKE_PTR_ARGS(OutputFileHandler, p_handler, ("TestLatticeBasedAngiogenesisLiteratePaper"));
RandomNumberGenerator::Instance()->Reseed(12345);
units::quantity<unit::length> reference_length(1.0 * unit::microns);
units::quantity<unit::time> reference_time(1.0* unit::hours);
BaseUnits::Instance()->SetReferenceLengthScale(reference_length);
BaseUnits::Instance()->SetReferenceTimeScale(reference_time);
boost::shared_ptr<RegularGrid<2> > p_grid = RegularGrid<2>::Create();
units::quantity<unit::length> grid_spacing = Owen11Parameters::mpLatticeSpacing->GetValue("User");
p_grid->SetSpacing(grid_spacing);
std::vector<unsigned> extents(3, 1);
extents[0] = 51; // num x
extents[1] = 51; // num_y
p_grid->SetExtents(extents);
p_grid->Write(p_handler);
boost::shared_ptr<VesselNode<2> > p_node11 = VesselNode<2>::Create(0.0, 400.0, 0.0, reference_length);
boost::shared_ptr<VesselNode<2> > p_node12 = VesselNode<2>::Create(2000.0, 400.0, 0.0, reference_length);
p_node11->GetFlowProperties()->SetIsInputNode(true);
p_node11->GetFlowProperties()->SetPressure(Owen11Parameters::mpInletPressure->GetValue("User"));
p_node12->GetFlowProperties()->SetIsOutputNode(true);
p_node12->GetFlowProperties()->SetPressure(Owen11Parameters::mpOutletPressure->GetValue("User"));
boost::shared_ptr<VesselNode<2> > p_node13 = VesselNode<2>::Create(2000.0, 1600.0, 0.0, reference_length);
boost::shared_ptr<VesselNode<2> > p_node14 = VesselNode<2>::Create(0.0, 1600.0, 0.0, reference_length);
p_node13->GetFlowProperties()->SetIsInputNode(true);
p_node13->GetFlowProperties()->SetPressure(Owen11Parameters::mpInletPressure->GetValue("User"));
p_node14->GetFlowProperties()->SetIsOutputNode(true);
p_node14->GetFlowProperties()->SetPressure(Owen11Parameters::mpOutletPressure->GetValue("User"));
boost::shared_ptr<Vessel<2> > p_vessel1 = Vessel<2>::Create(p_node11, p_node12);
boost::shared_ptr<Vessel<2> > p_vessel2 = Vessel<2>::Create(p_node13, p_node14);
boost::shared_ptr<VesselNetwork<2> > p_network = VesselNetwork<2>::Create();
p_network->AddVessel(p_vessel1);
p_network->AddVessel(p_vessel2);
p_network->Write(p_handler->GetOutputDirectoryFullPath() + "initial_network.vtp");
boost::shared_ptr<Owen11CellPopulationGenerator<2> > p_cell_population_genenerator = Owen11CellPopulationGenerator<2>::Create();
p_cell_population_genenerator->SetRegularGrid(p_grid);
p_cell_population_genenerator->SetVesselNetwork(p_network);
units::quantity<unit::length> tumour_radius(300.0 * unit::microns);
p_cell_population_genenerator->SetTumourRadius(tumour_radius);
boost::shared_ptr<CaBasedCellPopulation<2> > p_cell_population = p_cell_population_genenerator->Update();
boost::shared_ptr<LinearSteadyStateDiffusionReactionPde<2> > p_oxygen_pde = LinearSteadyStateDiffusionReactionPde<2>::Create();
p_oxygen_pde->SetIsotropicDiffusionConstant(Owen11Parameters::mpOxygenDiffusivity->GetValue("User"));
boost::shared_ptr<CellBasedDiscreteSource<2> > p_cell_oxygen_sink = CellBasedDiscreteSource<2>::Create();
p_cell_oxygen_sink->SetLinearInUConsumptionRatePerCell(Owen11Parameters::mpCellOxygenConsumptionRate->GetValue("User"));
p_oxygen_pde->AddDiscreteSource(p_cell_oxygen_sink);
boost::shared_ptr<VesselBasedDiscreteSource<2> > p_vessel_oxygen_source = VesselBasedDiscreteSource<2>::Create();
units::quantity<unit::solubility> oxygen_solubility_at_stp = Secomb04Parameters::mpOxygenVolumetricSolubility->GetValue("User") *
GenericParameters::mpGasConcentrationAtStp->GetValue("User");
units::quantity<unit::concentration> vessel_oxygen_concentration = oxygen_solubility_at_stp *
Owen11Parameters::mpReferencePartialPressure->GetValue("User");
p_vessel_oxygen_source->SetReferenceConcentration(vessel_oxygen_concentration);
p_vessel_oxygen_source->SetVesselPermeability(Owen11Parameters::mpVesselOxygenPermeability->GetValue("User"));
p_vessel_oxygen_source->SetReferenceHaematocrit(Owen11Parameters::mpInflowHaematocrit->GetValue("User"));
p_oxygen_pde->AddDiscreteSource(p_vessel_oxygen_source);
boost::shared_ptr<FiniteDifferenceSolver<2> > p_oxygen_solver = FiniteDifferenceSolver<2>::Create();
p_oxygen_solver->SetPde(p_oxygen_pde);
p_oxygen_solver->SetLabel("oxygen");
p_oxygen_solver->SetGrid(p_grid);
boost::shared_ptr<LinearSteadyStateDiffusionReactionPde<2> > p_vegf_pde = LinearSteadyStateDiffusionReactionPde<2>::Create();
p_vegf_pde->SetIsotropicDiffusionConstant(Owen11Parameters::mpVegfDiffusivity->GetValue("User"));
p_vegf_pde->SetContinuumLinearInUTerm(-Owen11Parameters::mpVegfDecayRate->GetValue("User"));
boost::shared_ptr<CellStateDependentDiscreteSource<2> > p_normal_and_quiescent_cell_source = CellStateDependentDiscreteSource<2>::Create();
std::map<unsigned, units::quantity<unit::concentration_flow_rate> > normal_and_quiescent_cell_rates;
std::map<unsigned, units::quantity<unit::concentration> > normal_and_quiescent_cell_rate_thresholds;
MAKE_PTR(QuiescentCancerCellMutationState, p_quiescent_cancer_state);
MAKE_PTR(WildTypeCellMutationState, p_normal_cell_state);
normal_and_quiescent_cell_rates[p_normal_cell_state->GetColour()] = Owen11Parameters::mpCellVegfSecretionRate->GetValue("User");
normal_and_quiescent_cell_rate_thresholds[p_normal_cell_state->GetColour()] = 0.27*unit::mole_per_metre_cubed;
normal_and_quiescent_cell_rates[p_quiescent_cancer_state->GetColour()] = Owen11Parameters::mpCellVegfSecretionRate->GetValue("User");
normal_and_quiescent_cell_rate_thresholds[p_quiescent_cancer_state->GetColour()] = 0.0*unit::mole_per_metre_cubed;
p_normal_and_quiescent_cell_source->SetStateRateMap(normal_and_quiescent_cell_rates);
p_normal_and_quiescent_cell_source->SetLabelName("VEGF");
p_normal_and_quiescent_cell_source->SetStateRateThresholdMap(normal_and_quiescent_cell_rate_thresholds);
p_vegf_pde->AddDiscreteSource(p_normal_and_quiescent_cell_source);
boost::shared_ptr<VesselBasedDiscreteSource<2> > p_vessel_vegf_sink = VesselBasedDiscreteSource<2>::Create();
p_vessel_vegf_sink->SetReferenceConcentration(0.0*unit::mole_per_metre_cubed);
p_vessel_vegf_sink->SetVesselPermeability(Owen11Parameters::mpVesselVegfPermeability->GetValue("User"));
p_vegf_pde->AddDiscreteSource(p_vessel_vegf_sink);
boost::shared_ptr<FiniteDifferenceSolver<2> > p_vegf_solver = FiniteDifferenceSolver<2>::Create();
p_vegf_solver->SetPde(p_vegf_pde);
p_vegf_solver->SetLabel("VEGF_Extracellular");
p_vegf_solver->SetGrid(p_grid);
units::quantity<unit::length> large_vessel_radius(25.0 * unit::microns);
p_network->SetSegmentRadii(large_vessel_radius);
units::quantity<unit::dynamic_viscosity> viscosity = Owen11Parameters::mpPlasmaViscosity->GetValue("User");
p_network->SetSegmentViscosity(viscosity);
boost::shared_ptr<VesselImpedanceCalculator<2> > p_impedance_calculator = VesselImpedanceCalculator<2>::Create();
boost::shared_ptr<ConstantHaematocritSolver<2> > p_haematocrit_calculator = ConstantHaematocritSolver<2>::Create();
p_haematocrit_calculator->SetHaematocrit(Owen11Parameters::mpInflowHaematocrit->GetValue("User"));
boost::shared_ptr<WallShearStressCalculator<2> > p_wss_calculator = WallShearStressCalculator<2>::Create();
boost::shared_ptr<MechanicalStimulusCalculator<2> > p_mech_stimulus_calculator = MechanicalStimulusCalculator<2>::Create();
boost::shared_ptr<MetabolicStimulusCalculator<2> > p_metabolic_stim_calculator = MetabolicStimulusCalculator<2>::Create();
boost::shared_ptr<ShrinkingStimulusCalculator<2> > p_shrinking_stimulus_calculator = ShrinkingStimulusCalculator<2>::Create();
boost::shared_ptr<ViscosityCalculator<2> > p_viscosity_calculator = ViscosityCalculator<2>::Create();
boost::shared_ptr<StructuralAdaptationSolver<2> > p_structural_adaptation_solver = StructuralAdaptationSolver<2>::Create();
p_structural_adaptation_solver->SetTolerance(0.0001);
p_structural_adaptation_solver->SetMaxIterations(100);
p_structural_adaptation_solver->SetTimeIncrement(Owen11Parameters::mpVesselRadiusUpdateTimestep->GetValue("User"));
p_structural_adaptation_solver->AddPreFlowSolveCalculator(p_impedance_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_haematocrit_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_wss_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_metabolic_stim_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_mech_stimulus_calculator);
// p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_shrinking_stimulus_calculator);
p_structural_adaptation_solver->AddPostFlowSolveCalculator(p_viscosity_calculator);
boost::shared_ptr<WallShearStressBasedRegressionSolver<2> > p_regression_solver =
WallShearStressBasedRegressionSolver<2>::Create();
boost::shared_ptr<AngiogenesisSolver<2> > p_angiogenesis_solver = AngiogenesisSolver<2>::Create();
boost::shared_ptr<Owen2011SproutingRule<2> > p_sprouting_rule = Owen2011SproutingRule<2>::Create();
boost::shared_ptr<Owen2011MigrationRule<2> > p_migration_rule = Owen2011MigrationRule<2>::Create();
p_angiogenesis_solver->SetMigrationRule(p_migration_rule);
p_angiogenesis_solver->SetSproutingRule(p_sprouting_rule);
p_sprouting_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_migration_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_angiogenesis_solver->SetVesselGrid(p_grid);
p_angiogenesis_solver->SetVesselNetwork(p_network);
boost::shared_ptr<MicrovesselSolver<2> > p_microvessel_solver = MicrovesselSolver<2>::Create();
p_microvessel_solver->SetVesselNetwork(p_network);
p_microvessel_solver->SetOutputFrequency(5);
p_microvessel_solver->AddDiscreteContinuumSolver(p_oxygen_solver);
p_microvessel_solver->AddDiscreteContinuumSolver(p_vegf_solver);
p_microvessel_solver->SetStructuralAdaptationSolver(p_structural_adaptation_solver);
p_microvessel_solver->SetRegressionSolver(p_regression_solver);
p_microvessel_solver->SetAngiogenesisSolver(p_angiogenesis_solver);
boost::shared_ptr<MicrovesselSimulationModifier<2> > p_microvessel_modifier = MicrovesselSimulationModifier<2>::Create();
p_microvessel_modifier->SetMicrovesselSolver(p_microvessel_solver);
std::vector<std::string> update_labels;
update_labels.push_back("oxygen");
update_labels.push_back("VEGF_Extracellular");
p_microvessel_modifier->SetCellDataUpdateLabels(update_labels);
OnLatticeSimulation<2> simulator(*p_cell_population);
simulator.AddSimulationModifier(p_microvessel_modifier);
boost::shared_ptr<ApoptoticCellKiller<2> > p_apoptotic_cell_killer(new ApoptoticCellKiller<2>(p_cell_population.get()));
simulator.AddCellKiller(p_apoptotic_cell_killer);
boost::shared_ptr<Owen2011TrackingModifier<2> > p_owen11_tracking_modifier(new Owen2011TrackingModifier<2>);
simulator.AddSimulationModifier(p_owen11_tracking_modifier);
simulator.SetOutputDirectory("TestLatticeBasedAngiogenesisLiteratePaper");
simulator.SetSamplingTimestepMultiple(5);
simulator.SetDt(0.5);
simulator.SetEndTime(20.0);
simulator.Solve();
ParameterCollection::Instance()->DumpToFile(p_handler->GetOutputDirectoryFullPath()+"parameter_collection.xml");
}
};