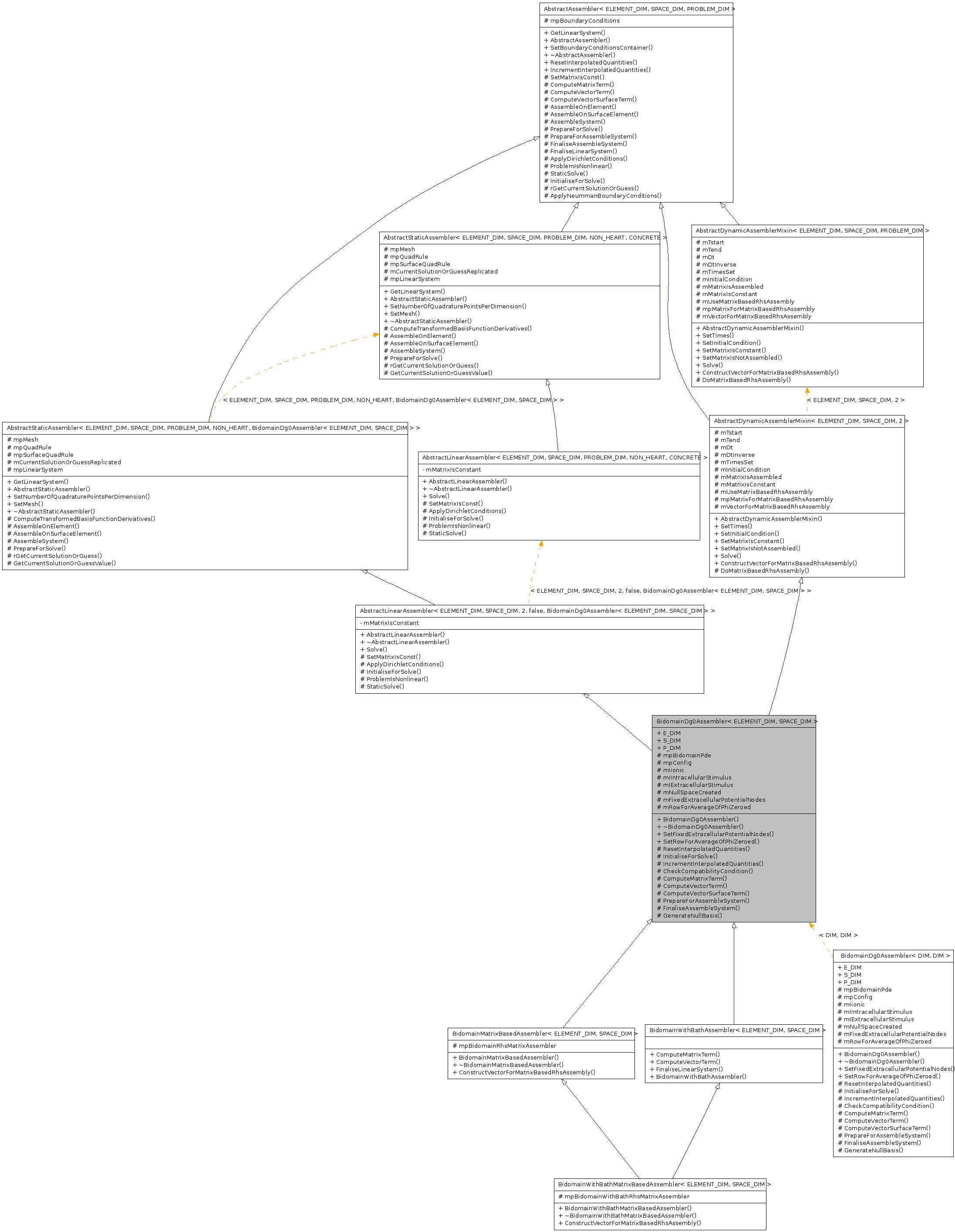
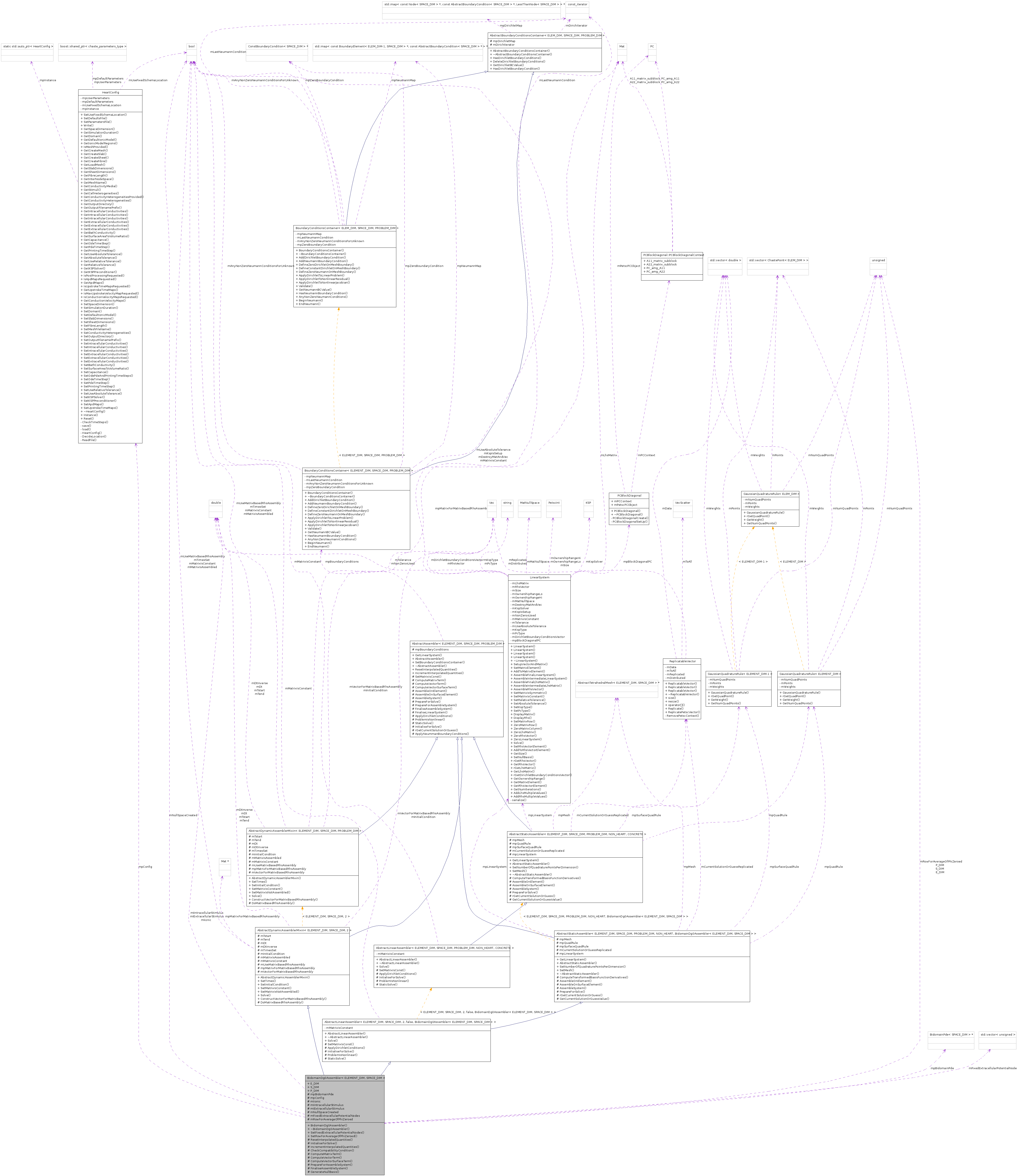
BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > Class Template Reference
#include <BidomainDg0Assembler.hpp>
Public Member Functions | |
| BidomainDg0Assembler (AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > *pMesh, BidomainPde< SPACE_DIM > *pPde, BoundaryConditionsContainer< ELEMENT_DIM, SPACE_DIM, 2 > *pBcc, unsigned numQuadPoints=2) | |
| ~BidomainDg0Assembler () | |
| void | SetFixedExtracellularPotentialNodes (std::vector< unsigned > fixedExtracellularPotentialNodes) |
| void | SetRowForAverageOfPhiZeroed (unsigned rowMeanPhiEZero) |
Static Public Attributes | |
| static const unsigned | E_DIM = ELEMENT_DIM |
| static const unsigned | S_DIM = SPACE_DIM |
| static const unsigned | P_DIM = 2u |
Protected Types | |
| typedef BidomainDg0Assembler < ELEMENT_DIM, SPACE_DIM > | SelfType |
| typedef AbstractLinearAssembler < ELEMENT_DIM, SPACE_DIM, 2, false, SelfType > | BaseClassType |
Protected Member Functions | |
| void | ResetInterpolatedQuantities () |
| void | InitialiseForSolve (Vec initialSolution) |
| void | IncrementInterpolatedQuantities (double phiI, const Node< SPACE_DIM > *pNode) |
| virtual void | CheckCompatibilityCondition () |
| virtual c_matrix< double, 2 *(ELEMENT_DIM+1), 2 *(ELEMENT_DIM+1)> | ComputeMatrixTerm (c_vector< double, ELEMENT_DIM+1 > &rPhi, c_matrix< double, ELEMENT_DIM, ELEMENT_DIM+1 > &rGradPhi, ChastePoint< SPACE_DIM > &rX, c_vector< double, 2 > &rU, c_matrix< double, 2, SPACE_DIM > &rGradU, Element< ELEMENT_DIM, SPACE_DIM > *pElement) |
| virtual c_vector< double, 2 *(ELEMENT_DIM+1)> | ComputeVectorTerm (c_vector< double, ELEMENT_DIM+1 > &rPhi, c_matrix< double, ELEMENT_DIM, ELEMENT_DIM+1 > &rGradPhi, ChastePoint< SPACE_DIM > &rX, c_vector< double, 2 > &u, c_matrix< double, 2, SPACE_DIM > &rGradU, Element< ELEMENT_DIM, SPACE_DIM > *pElement) |
| virtual c_vector< double, 2 *ELEMENT_DIM > | ComputeVectorSurfaceTerm (const BoundaryElement< ELEMENT_DIM-1, SPACE_DIM > &rSurfaceElement, c_vector< double, ELEMENT_DIM > &rPhi, ChastePoint< SPACE_DIM > &rX) |
| virtual void | PrepareForAssembleSystem (Vec existingSolution, double time) |
| virtual void | FinaliseAssembleSystem (Vec existingSolution, double time) |
| virtual Vec | GenerateNullBasis () const |
Protected Attributes | |
| BidomainPde< SPACE_DIM > * | mpBidomainPde |
| HeartConfig * | mpConfig |
| double | mIionic |
| double | mIIntracellularStimulus |
| double | mIExtracellularStimulus |
| bool | mNullSpaceCreated |
| std::vector< unsigned > | mFixedExtracellularPotentialNodes |
| unsigned | mRowForAverageOfPhiZeroed |
Friends | |
| class | AbstractStaticAssembler< ELEMENT_DIM, SPACE_DIM, 2, false, SelfType > |
| Allow the AbstractStaticAssembler to call our private/protected methods using static polymorphism. | |
Detailed Description
template<unsigned ELEMENT_DIM, unsigned SPACE_DIM>
class BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >
BidomainDg0AssemblerThe 2 unknowns are voltage and extracellular potential.
This assembler interpolates quantities such as ionic currents and stimuli from their nodal values (obtained from a BidomainPde) onto a gauss point, and uses the interpolated values in assembly. The assembler also creates boundary conditions, which are zero-Neumann boundary conditions on the surface unless SetFixedExtracellularPotentialNodes() is called.
The user should call Solve() from the superclass AbstractDynamicAssemblerMixin.
NOTE: if any cells have a non-zero extracellular stimulus, phi_e must be fixed at some nodes (using SetFixedExtracellularPotentialNodes() ), else no solution is possible.
Definition at line 68 of file BidomainDg0Assembler.hpp.
Member Typedef Documentation
typedef BidomainDg0Assembler<ELEMENT_DIM, SPACE_DIM> BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::SelfType [protected] |
This type (to save typing).
Definition at line 79 of file BidomainDg0Assembler.hpp.
typedef AbstractLinearAssembler<ELEMENT_DIM, SPACE_DIM, 2, false, SelfType> BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::BaseClassType [protected] |
Base class type (to save typing).
Definition at line 80 of file BidomainDg0Assembler.hpp.
Constructor & Destructor Documentation
| BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::BidomainDg0Assembler | ( | AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > * | pMesh, | |
| BidomainPde< SPACE_DIM > * | pPde, | |||
| BoundaryConditionsContainer< ELEMENT_DIM, SPACE_DIM, 2 > * | pBcc, | |||
| unsigned | numQuadPoints = 2 | |||
| ) | [inline] |
Constructor stores the mesh and pde and sets up boundary conditions.
- Parameters:
-
pMesh pointer to the mesh pPde pointer to the PDE pBcc pointer to the boundary conditions numQuadPoints number of quadrature points (defaults to 2)
Definition at line 323 of file BidomainDg0Assembler.cpp.
References HeartConfig::Instance(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mNullSpaceCreated, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpBidomainPde, AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpBoundaryConditions, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpConfig, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mRowForAverageOfPhiZeroed, AbstractDynamicAssemblerMixin< ELEMENT_DIM, SPACE_DIM, 2 >::SetMatrixIsConstant(), and AbstractStaticAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM, NON_HEART, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > >::SetMesh().
| BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::~BidomainDg0Assembler | ( | ) | [inline] |
Destructor.
Definition at line 351 of file BidomainDg0Assembler.cpp.
Member Function Documentation
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ResetInterpolatedQuantities | ( | void | ) | [inline, protected, virtual] |
Overridden ResetInterpolatedQuantities() method.
Reimplemented from AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >.
Definition at line 37 of file BidomainDg0Assembler.cpp.
References BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIIntracellularStimulus, and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIionic.
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::InitialiseForSolve | ( | Vec | initialSolution | ) | [inline, protected, virtual] |
Create the linear system object if it hasn't been already. Can use an initial solution as PETSc template, or base it on the mesh size.
- Parameters:
-
initialSolution an initial guess
Reimplemented from AbstractLinearAssembler< ELEMENT_DIM, SPACE_DIM, 2, false, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > >.
Definition at line 44 of file BidomainDg0Assembler.cpp.
References HeartConfig::GetAbsoluteTolerance(), HeartConfig::GetRelativeTolerance(), AbstractLinearAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM, NON_HEART, CONCRETE >::InitialiseForSolve(), HeartConfig::Instance(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpConfig, AbstractStaticAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM, NON_HEART, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > >::mpLinearSystem, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mRowForAverageOfPhiZeroed, LinearSystem::SetAbsoluteTolerance(), LinearSystem::SetKspType(), LinearSystem::SetMatrixIsSymmetric(), LinearSystem::SetPcType(), and LinearSystem::SetRelativeTolerance().
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::IncrementInterpolatedQuantities | ( | double | phiI, | |
| const Node< SPACE_DIM > * | pNode | |||
| ) | [inline, protected, virtual] |
Overridden IncrementInterpolatedQuantities() method.
- Parameters:
-
phiI pNode
Reimplemented from AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >.
Definition at line 86 of file BidomainDg0Assembler.cpp.
References Node< SPACE_DIM >::GetIndex(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIIntracellularStimulus, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIionic, and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpBidomainPde.
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::CheckCompatibilityCondition | ( | ) | [inline, protected, virtual] |
Checks whether the linear system will have a solution (if so, infinite solutions) instead of zero solutions. The condition is, if the linear system is Ax=b, that sum b_i over for all the PHI_E components (ie i=1,3,5,..) is zero.
This check is not made if running in parallel, or in debug mode.
The reason why the sum must be zero: the Fredholm alternative states that a singular system Ax=b has a solution if and only if v.b=0 for all v in ker(A) (ie all v such that Av=b). The nullspace ker(A) is one dimensional with basis vector v = (0,1,0,1....,0,1), so v.b = sum_{i=1,3,5..} b_i.
Definition at line 288 of file BidomainDg0Assembler.cpp.
References PetscTools::GetNumProcs(), AbstractBoundaryConditionsContainer< ELEM_DIM, SPACE_DIM, PROBLEM_DIM >::HasDirichletBoundaryConditions(), AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpBoundaryConditions, AbstractStaticAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM, NON_HEART, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > >::mpLinearSystem, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mRowForAverageOfPhiZeroed, and LinearSystem::rGetRhsVector().
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::FinaliseAssembleSystem().
| c_matrix< double, 2 *(ELEMENT_DIM+1), 2 *(ELEMENT_DIM+1)> BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeMatrixTerm | ( | c_vector< double, ELEMENT_DIM+1 > & | rPhi, | |
| c_matrix< double, ELEMENT_DIM, ELEMENT_DIM+1 > & | rGradPhi, | |||
| ChastePoint< SPACE_DIM > & | rX, | |||
| c_vector< double, 2 > & | rU, | |||
| c_matrix< double, 2, SPACE_DIM > & | rGradU, | |||
| Element< ELEMENT_DIM, SPACE_DIM > * | pElement | |||
| ) | [inline, protected, virtual] |
This method is called by AssembleOnElement() and tells the assembler the contribution to add to the element stiffness matrix.
- Parameters:
-
rPhi The basis functions, rPhi(i) = phi_i, i=1..numBases rGradPhi Basis gradients, rGradPhi(i,j) = d(phi_j)/d(X_i) rX The point in space rU The unknown as a vector, u(i) = u_i rGradU The gradient of the unknown as a matrix, rGradU(i,j) = d(u_i)/d(X_j) pElement Pointer to the element
Reimplemented in BidomainWithBathAssembler< ELEMENT_DIM, SPACE_DIM >.
Definition at line 96 of file BidomainDg0Assembler.cpp.
References HeartConfig::GetCapacitance(), AbstractElement< ELEMENT_DIM, SPACE_DIM >::GetIndex(), HeartConfig::GetSurfaceAreaToVolumeRatio(), AbstractDynamicAssemblerMixin< ELEMENT_DIM, SPACE_DIM, 2 >::mDt, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpBidomainPde, and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpConfig.
Referenced by BidomainWithBathAssembler< ELEMENT_DIM, SPACE_DIM >::ComputeMatrixTerm().
| c_vector< double, 2 *(ELEMENT_DIM+1)> BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeVectorTerm | ( | c_vector< double, ELEMENT_DIM+1 > & | rPhi, | |
| c_matrix< double, ELEMENT_DIM, ELEMENT_DIM+1 > & | rGradPhi, | |||
| ChastePoint< SPACE_DIM > & | rX, | |||
| c_vector< double, 2 > & | u, | |||
| c_matrix< double, 2, SPACE_DIM > & | rGradU, | |||
| Element< ELEMENT_DIM, SPACE_DIM > * | pElement | |||
| ) | [inline, protected, virtual] |
This method is called by AssembleOnElement() and tells the assembler the contribution to add to the element stiffness vector.
- Parameters:
-
rPhi The basis functions, rPhi(i) = phi_i, i=1..numBases rGradPhi Basis gradients, rGradPhi(i,j) = d(phi_j)/d(X_i) rX The point in space u The unknown as a vector, u(i) = u_i rGradU The gradient of the unknown as a matrix, rGradU(i,j) = d(u_i)/d(X_j) pElement Pointer to the element
Reimplemented in BidomainWithBathAssembler< ELEMENT_DIM, SPACE_DIM >.
Definition at line 152 of file BidomainDg0Assembler.cpp.
References HeartConfig::GetCapacitance(), HeartConfig::GetSurfaceAreaToVolumeRatio(), AbstractDynamicAssemblerMixin< ELEMENT_DIM, SPACE_DIM, 2 >::mDt, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIIntracellularStimulus, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIionic, and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpConfig.
Referenced by BidomainWithBathAssembler< ELEMENT_DIM, SPACE_DIM >::ComputeVectorTerm().
| c_vector< double, 2 *ELEMENT_DIM > BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeVectorSurfaceTerm | ( | const BoundaryElement< ELEMENT_DIM-1, SPACE_DIM > & | rSurfaceElement, | |
| c_vector< double, ELEMENT_DIM > & | rPhi, | |||
| ChastePoint< SPACE_DIM > & | rX | |||
| ) | [inline, protected, virtual] |
This method returns the vector to be added to element stiffness vector for a given gauss point in BoundaryElement. The arguments are the bases, x and current solution computed at the Gauss point. The returned vector will be multiplied by the gauss weight and jacobian determinent and added to the element stiffness matrix (see AssembleOnElement()).
--This method has to be implemented in the concrete class--
- Parameters:
-
rSurfaceElement the element which is being considered. rPhi The basis functions, rPhi(i) = phi_i, i=1..numBases rX The point in space
Implements AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >.
Definition at line 181 of file BidomainDg0Assembler.cpp.
References BoundaryConditionsContainer< ELEM_DIM, SPACE_DIM, PROBLEM_DIM >::GetNeumannBCValue(), and AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpBoundaryConditions.
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::PrepareForAssembleSystem | ( | Vec | existingSolution, | |
| double | time | |||
| ) | [inline, protected, virtual] |
PrepareForAssembleSystem
Called at the beginning of AbstractLinearAssembler::AssembleSystem() after the system. Here, used to integrate cell odes.
- Parameters:
-
existingSolution is the voltage to feed into the cell models time the simulation time
Reimplemented from AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >.
Definition at line 203 of file BidomainDg0Assembler.cpp.
References AbstractDynamicAssemblerMixin< ELEMENT_DIM, SPACE_DIM, 2 >::mDt, and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpBidomainPde.
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::FinaliseAssembleSystem | ( | Vec | existingSolution, | |
| double | time | |||
| ) | [inline, protected, virtual] |
FinaliseAssembleSystem
Called by AbstractLinearAssmebler::AssembleSystem() after the system has been assembled. Here, used to avoid problems with phi_e drifting by one of 3 methods: pinning nodes, using a null space, or using an "average phi_e = 0" row.
- Parameters:
-
existingSolution voltages (phi_e may need shifting) time
Reimplemented from AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >.
Definition at line 233 of file BidomainDg0Assembler.cpp.
References LinearSystem::AssembleFinalLhsMatrix(), LinearSystem::AssembleRhsVector(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::CheckCompatibilityCondition(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::GenerateNullBasis(), LinearSystem::GetSize(), AbstractBoundaryConditionsContainer< ELEM_DIM, SPACE_DIM, PROBLEM_DIM >::HasDirichletBoundaryConditions(), AbstractDynamicAssemblerMixin< ELEMENT_DIM, SPACE_DIM, 2 >::mMatrixIsAssembled, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mNullSpaceCreated, AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpBoundaryConditions, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpConfig, AbstractStaticAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM, NON_HEART, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > >::mpLinearSystem, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mRowForAverageOfPhiZeroed, HeartConfig::SetKSPSolver(), LinearSystem::SetKspType(), LinearSystem::SetMatrixElement(), LinearSystem::SetNullBasis(), LinearSystem::SetRhsVectorElement(), and LinearSystem::ZeroMatrixRow().
| Vec BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::GenerateNullBasis | ( | ) | const [inline, protected, virtual] |
GenerateNullBasis
Called by FinaliseAssembleSystem to get the null basis to use for the particular formulation of the bidomain equations used. Parabolic-Elliptic in BidomainDg0Assembler.
Definition at line 209 of file BidomainDg0Assembler.cpp.
References DistributedVector::Begin(), DistributedVectorFactory::CreateDistributedVector(), DistributedVectorFactory::CreateVec(), DistributedVector::End(), AbstractStaticAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM, NON_HEART, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > >::mpMesh, and DistributedVector::Restore().
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::FinaliseAssembleSystem().
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::SetFixedExtracellularPotentialNodes | ( | std::vector< unsigned > | fixedExtracellularPotentialNodes | ) | [inline] |
Set the nodes at which phi_e (the extracellular potential) is fixed to zero. This does not necessarily have to be called. If it is not, phi_e is only defined up to a constant.
- Parameters:
-
fixedExtracellularPotentialNodes the nodes to be fixed.
Definition at line 356 of file BidomainDg0Assembler.cpp.
References BoundaryConditionsContainer< ELEM_DIM, SPACE_DIM, PROBLEM_DIM >::AddDirichletBoundaryCondition(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mFixedExtracellularPotentialNodes, AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpBoundaryConditions, and AbstractStaticAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM, NON_HEART, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM > >::mpMesh.
Referenced by BidomainProblem< DIM >::CreateAssembler().
| void BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::SetRowForAverageOfPhiZeroed | ( | unsigned | rowMeanPhiEZero | ) | [inline] |
Used when removing a single row to resolve singularity and replacing it with a constraint on the average phi_e being zero. It is set from the problem class.
- Parameters:
-
rowMeanPhiEZero indicates the row of the matrix to be replaced. Ought to be an odd number...
Definition at line 381 of file BidomainDg0Assembler.cpp.
References BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mRowForAverageOfPhiZeroed.
Referenced by BidomainProblem< DIM >::CreateAssembler().
Member Data Documentation
const unsigned BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::E_DIM = ELEMENT_DIM [static] |
The element dimension (to save typing).
Definition at line 73 of file BidomainDg0Assembler.hpp.
const unsigned BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::S_DIM = SPACE_DIM [static] |
The space dimension (to save typing).
Definition at line 74 of file BidomainDg0Assembler.hpp.
const unsigned BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::P_DIM = 2u [static] |
The problem dimension (to save typing).
Definition at line 75 of file BidomainDg0Assembler.hpp.
BidomainPde<SPACE_DIM>* BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpBidomainPde [protected] |
The PDE to be solved.
Definition at line 86 of file BidomainDg0Assembler.hpp.
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::BidomainDg0Assembler(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeMatrixTerm(), BidomainWithBathMatrixBasedAssembler< ELEMENT_DIM, SPACE_DIM >::ConstructVectorForMatrixBasedRhsAssembly(), BidomainMatrixBasedAssembler< ELEMENT_DIM, SPACE_DIM >::ConstructVectorForMatrixBasedRhsAssembly(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::IncrementInterpolatedQuantities(), and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::PrepareForAssembleSystem().
HeartConfig* BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mpConfig [protected] |
Local cache of the configuration singleton instance
Definition at line 89 of file BidomainDg0Assembler.hpp.
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::BidomainDg0Assembler(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeMatrixTerm(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeVectorTerm(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::FinaliseAssembleSystem(), BidomainWithBathAssembler< ELEMENT_DIM, SPACE_DIM >::FinaliseLinearSystem(), and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::InitialiseForSolve().
double BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIionic [protected] |
Ionic current to be interpolated from cache
Definition at line 92 of file BidomainDg0Assembler.hpp.
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeVectorTerm(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::IncrementInterpolatedQuantities(), and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ResetInterpolatedQuantities().
double BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIIntracellularStimulus [protected] |
Intracellular stimulus to be interpolated from cache
Definition at line 94 of file BidomainDg0Assembler.hpp.
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeVectorTerm(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::IncrementInterpolatedQuantities(), and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ResetInterpolatedQuantities().
double BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mIExtracellularStimulus [protected] |
Extracellular stimulus to be interpolated from cache
Definition at line 96 of file BidomainDg0Assembler.hpp.
bool BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mNullSpaceCreated [protected] |
Used when intialising null-space solver to resolve singularity
Definition at line 99 of file BidomainDg0Assembler.hpp.
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::BidomainDg0Assembler(), and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::FinaliseAssembleSystem().
std::vector<unsigned> BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mFixedExtracellularPotentialNodes [protected] |
Used when pinning nodes to resolve singularity. This vector indicates the global indices of the nodes to be pinned
Definition at line 104 of file BidomainDg0Assembler.hpp.
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::SetFixedExtracellularPotentialNodes().
unsigned BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::mRowForAverageOfPhiZeroed [protected] |
Used when removing a single row to resolve singularity and replacing it with a constraint on the average phi_e being zero. This number indicates the row of the matrix to be replaced. This is INT_MAX if unset. It is set from the problem class.
Definition at line 111 of file BidomainDg0Assembler.hpp.
Referenced by BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::BidomainDg0Assembler(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::CheckCompatibilityCondition(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::FinaliseAssembleSystem(), BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::InitialiseForSolve(), and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::SetRowForAverageOfPhiZeroed().
The documentation for this class was generated from the following files:
- /tmp/release_1.1/heart/src/solver/BidomainDg0Assembler.hpp
- /tmp/release_1.1/heart/src/solver/BidomainDg0Assembler.cpp
 1.5.5
1.5.5