MonodomainPde< ELEM_DIM, SPACE_DIM > Class Template Reference
#include <MonodomainPde.hpp>
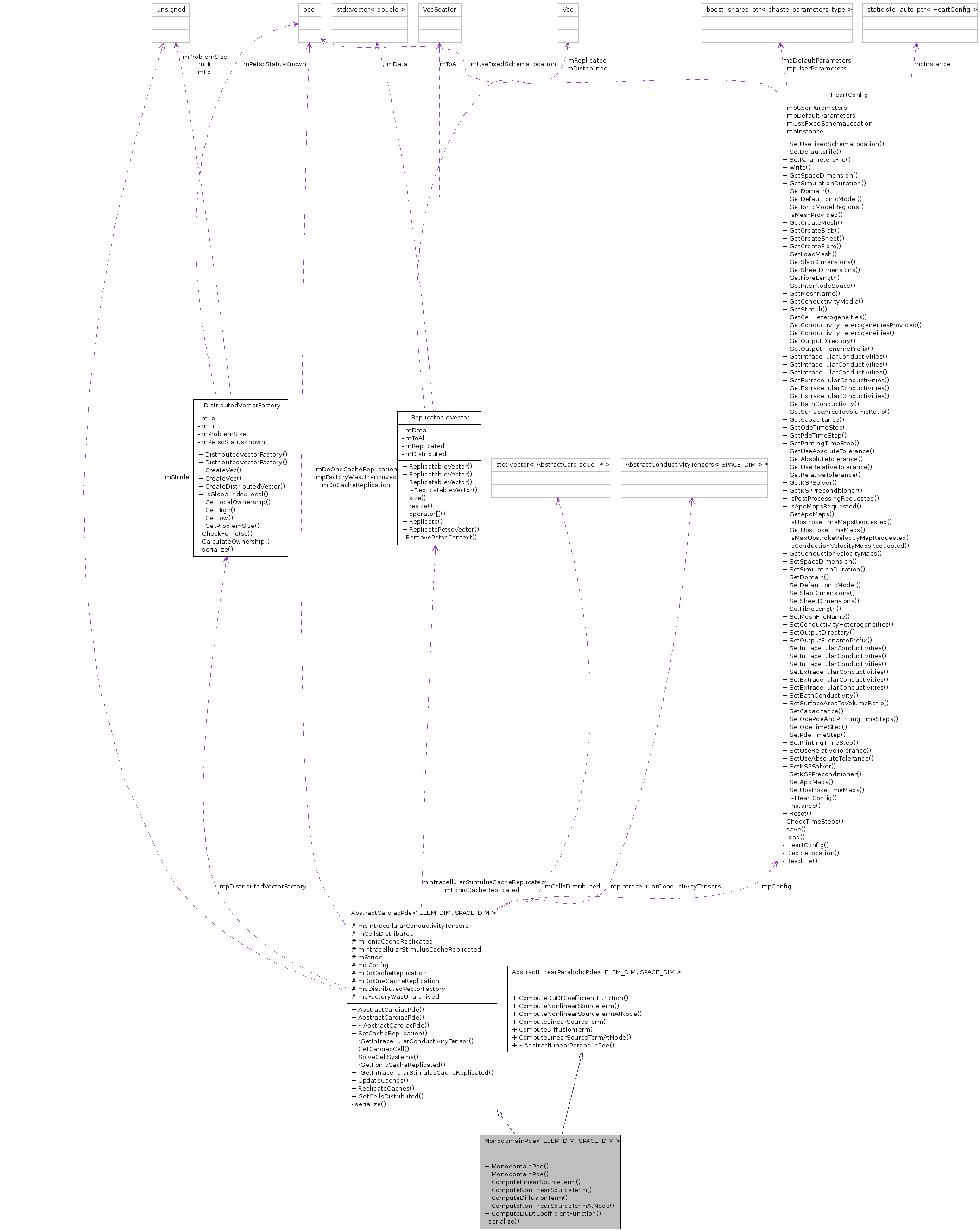
Public Member Functions | |
| MonodomainPde (AbstractCardiacCellFactory< ELEM_DIM, SPACE_DIM > *pCellFactory) | |
| Constructor. | |
| MonodomainPde (std::vector< AbstractCardiacCell * > &rCellsDistributed) | |
| double | ComputeLinearSourceTerm (const ChastePoint< SPACE_DIM > &) |
| double | ComputeNonlinearSourceTerm (const ChastePoint< SPACE_DIM > &, double) |
| virtual c_matrix< double, SPACE_DIM, SPACE_DIM > | ComputeDiffusionTerm (const ChastePoint< SPACE_DIM > &rX, Element< ELEM_DIM, SPACE_DIM > *pElement) |
| double | ComputeNonlinearSourceTermAtNode (const Node< SPACE_DIM > &node, double) |
| double | ComputeDuDtCoefficientFunction (const ChastePoint< SPACE_DIM > &) |
Private Member Functions | |
| template<class Archive> | |
| void | serialize (Archive &archive, const unsigned int version) |
Friends | |
| class | TestMonodomainPde |
| class | boost::serialization::access |
Detailed Description
template<unsigned ELEM_DIM, unsigned SPACE_DIM = ELEM_DIM>
class MonodomainPde< ELEM_DIM, SPACE_DIM >
MonodomainPde class.The monodomain equation is of the form: A (C dV/dt + Iionic) +Istim = Div( sigma_i Grad(V) )
where A is the surface area to volume ratio (1/cm), C is the capacitance (uF/cm^2), sigma_i is the intracellular conductivity (mS/cm), I_ionic is the ionic current (uA/cm^2), I_stim is the intracellular stimulus current (uA/cm^3).
Note that default values of A, C and sigma_i are stored in the parent class
Definition at line 62 of file MonodomainPde.hpp.
Constructor & Destructor Documentation
| MonodomainPde< ELEM_DIM, SPACE_DIM >::MonodomainPde | ( | std::vector< AbstractCardiacCell * > & | rCellsDistributed | ) | [inline] |
Another constructor (for archiving)
- Parameters:
-
rCellsDistributed local cell models (recovered from archive)
Definition at line 39 of file MonodomainPde.cpp.
Member Function Documentation
| void MonodomainPde< ELEM_DIM, SPACE_DIM >::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Archive the member variables.
- Parameters:
-
archive version
Reimplemented from AbstractCardiacPde< ELEM_DIM, SPACE_DIM >.
Definition at line 76 of file MonodomainPde.hpp.
| double MonodomainPde< ELEM_DIM, SPACE_DIM >::ComputeLinearSourceTerm | ( | const ChastePoint< SPACE_DIM > & | ) | [inline, virtual] |
This should not be called; use ComputeLinearSourceTermAtNode instead
Implements AbstractLinearParabolicPde< ELEM_DIM, SPACE_DIM >.
Definition at line 49 of file MonodomainPde.cpp.
| double MonodomainPde< ELEM_DIM, SPACE_DIM >::ComputeNonlinearSourceTerm | ( | const ChastePoint< SPACE_DIM > & | , | |
| double | ||||
| ) | [inline, virtual] |
This should not be called; use ComputeNonlinearSourceTermAtNode instead
Implements AbstractLinearParabolicPde< ELEM_DIM, SPACE_DIM >.
Definition at line 55 of file MonodomainPde.cpp.
| c_matrix< double, SPACE_DIM, SPACE_DIM > MonodomainPde< ELEM_DIM, SPACE_DIM >::ComputeDiffusionTerm | ( | const ChastePoint< SPACE_DIM > & | rX, | |
| Element< ELEM_DIM, SPACE_DIM > * | pElement | |||
| ) | [inline, virtual] |
Compute the diffusion term at a given point.
- Parameters:
-
rX The point in space at which the diffusion term is computed. pElement the element for which to compute the contribution
- Returns:
- A matrix.
Implements AbstractLinearParabolicPde< ELEM_DIM, SPACE_DIM >.
Definition at line 64 of file MonodomainPde.cpp.
References AbstractElement< ELEMENT_DIM, SPACE_DIM >::GetIndex(), and AbstractCardiacPde< ELEM_DIM, SPACE_DIM >::mpIntracellularConductivityTensors.
| double MonodomainPde< ELEM_DIM, SPACE_DIM >::ComputeNonlinearSourceTermAtNode | ( | const Node< SPACE_DIM > & | rNode, | |
| double | u | |||
| ) | [inline, virtual] |
Compute nonlinear source term at a node.
- Parameters:
-
rNode the node at which the nonlinear source term is computed u the value of the dependent variable at the node
Reimplemented from AbstractLinearParabolicPde< ELEM_DIM, SPACE_DIM >.
Definition at line 73 of file MonodomainPde.cpp.
References Node< SPACE_DIM >::GetIndex(), HeartConfig::GetSurfaceAreaToVolumeRatio(), AbstractCardiacPde< ELEM_DIM, SPACE_DIM >::mIionicCacheReplicated, AbstractCardiacPde< ELEM_DIM, SPACE_DIM >::mIntracellularStimulusCacheReplicated, and AbstractCardiacPde< ELEM_DIM, SPACE_DIM >::mpConfig.
Referenced by MonodomainMatrixBasedAssembler< ELEMENT_DIM, SPACE_DIM >::ConstructVectorForMatrixBasedRhsAssembly(), and MonodomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::IncrementInterpolatedQuantities().
| double MonodomainPde< ELEM_DIM, SPACE_DIM >::ComputeDuDtCoefficientFunction | ( | const ChastePoint< SPACE_DIM > & | rX | ) | [inline, virtual] |
The function c(x) in "c(x) du/dt = Grad.(DiffusionTerm(x)*Grad(u))+LinearSourceTerm(x)+NonlinearSourceTerm(x, u)"
- Parameters:
-
rX the point in space at which the function c is computed
Implements AbstractLinearParabolicPde< ELEM_DIM, SPACE_DIM >.
Definition at line 84 of file MonodomainPde.cpp.
References HeartConfig::GetCapacitance(), HeartConfig::GetSurfaceAreaToVolumeRatio(), and AbstractCardiacPde< ELEM_DIM, SPACE_DIM >::mpConfig.
Referenced by MonodomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::ComputeVectorTerm(), and MonodomainMatrixBasedAssembler< ELEMENT_DIM, SPACE_DIM >::ConstructVectorForMatrixBasedRhsAssembly().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented from AbstractCardiacPde< ELEM_DIM, SPACE_DIM >.
Definition at line 68 of file MonodomainPde.hpp.
The documentation for this class was generated from the following files:
- /tmp/release_1.1/heart/src/pdes/MonodomainPde.hpp
- /tmp/release_1.1/heart/src/pdes/MonodomainPde.cpp
 1.5.5
1.5.5