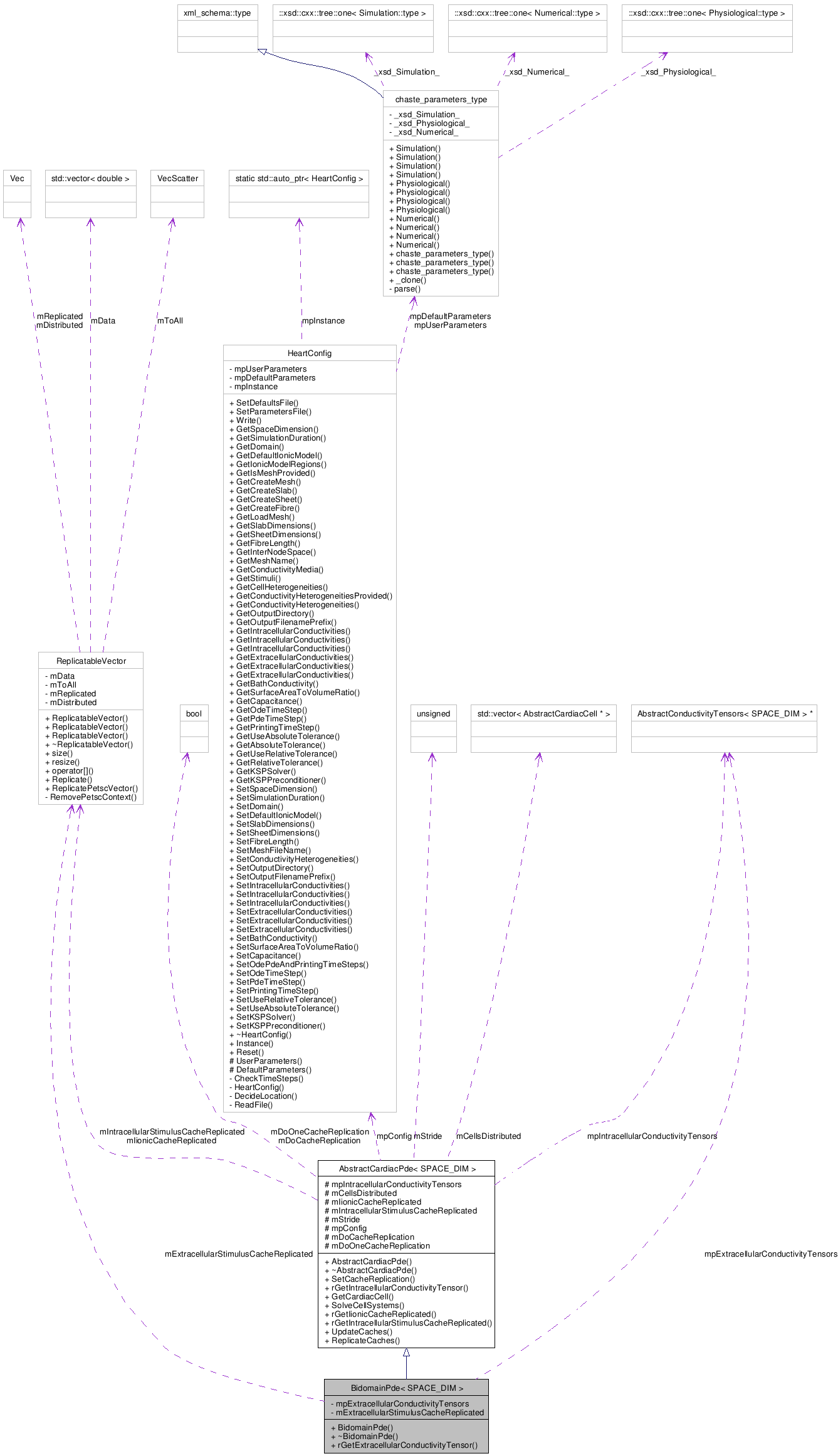
BidomainPde< SPACE_DIM > Class Template Reference
#include <BidomainPde.hpp>
Public Member Functions | |
| BidomainPde (AbstractCardiacCellFactory< SPACE_DIM > *pCellFactory) | |
|
const c_matrix< double, SPACE_DIM, SPACE_DIM > & | rGetExtracellularConductivityTensor (unsigned elementIndex) |
Private Attributes | |
|
AbstractConductivityTensors < SPACE_DIM > * | mpExtracellularConductivityTensors |
| ReplicatableVector | mExtracellularStimulusCacheReplicated |
Detailed Description
template<unsigned SPACE_DIM>
class BidomainPde< SPACE_DIM >
BidomainPde class.The bidmain equation is of the form:
A_m ( C_m d(V_m)/dt + I_ion ) = div ( sigma_i grad( V_m + phi_e ) ) + I_intra_stim and div ( (sigma_i + sigma_e) grad phi_e + sigma_i (grad V_m) ) + I_extra_stim
where V_m is the trans-membrane potential = phi_i - phi_e (mV), phi_i is the intracellular potential (mV), phi_e is the intracellular potential (mV), and A_m is the surface area to volume ratio of the cell membrane (1/cm), C_m is the membrane capacitance (uF/cm^2), sigma_i is the intracellular conductivity tensor (mS/cm), sigma_e is the intracellular conductivity tensor (mS/cm), and I_ion is the ionic current (uA/cm^2), I_intra_stim is the internal stimulus (uA/cm^3), I_extra_stim is the external stimulus (uA/cm^3).
Note: I_extra_stim can only be zero now. An extracellular stimulus can be applied as a boundary condition, in which case the units are uA/cm^2.
Definition at line 66 of file BidomainPde.hpp.
Constructor & Destructor Documentation
| BidomainPde< SPACE_DIM >::BidomainPde | ( | AbstractCardiacCellFactory< SPACE_DIM > * | pCellFactory | ) | [inline] |
Constructor sets up extracellular conductivity tensors.
Definition at line 39 of file BidomainPde.cpp.
References AbstractCardiacCellFactory< SPACE_DIM >::GetMesh(), and AbstractCardiacCellFactory< SPACE_DIM >::GetNumberOfCells().
The documentation for this class was generated from the following files:
- /tmp/release_1/heart/src/pdes/BidomainPde.hpp
- /tmp/release_1/heart/src/pdes/BidomainPde.cpp
 1.5.5
1.5.5