BidomainProblem< DIM > Class Template Reference
#include <BidomainProblem.hpp>

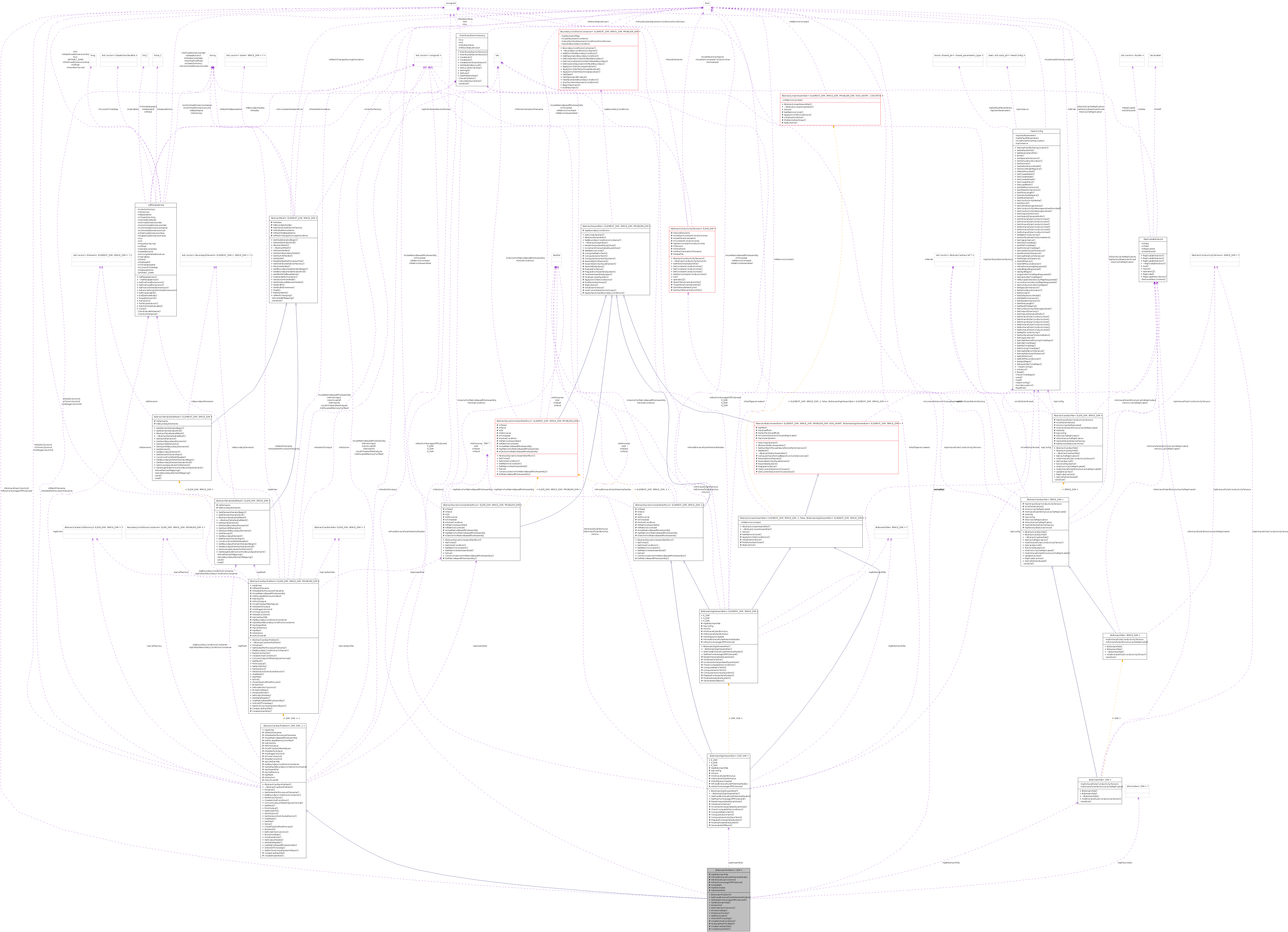
Public Member Functions | |
| BidomainProblem (AbstractCardiacCellFactory< DIM > *pCellFactory, bool hasBath=false) | |
| void | SetFixedExtracellularPotentialNodes (std::vector< unsigned > nodes) |
| void | SetNodeForAverageOfPhiZeroed (unsigned node) |
| BidomainPde< DIM > * | GetBidomainPde () |
| void | WriteInfo (double time) |
| virtual void | DefineWriterColumns () |
| virtual void | WriteOneStep (double time, Vec voltageVec) |
| void | PreSolveChecks () |
| void | SetElectrodes (Electrodes< DIM > &rElectrodes) |
| void | OnEndOfTimestep (double time) |
Protected Member Functions | |
| Vec | CreateInitialCondition () |
| void | AnalyseMeshForBath () |
| virtual AbstractCardiacPde< DIM > * | CreateCardiacPde () |
| virtual AbstractDynamicAssemblerMixin < DIM, DIM, 2 > * | CreateAssembler () |
Protected Attributes | |
| BidomainPde< DIM > * | mpBidomainPde |
| std::vector< unsigned > | mFixedExtracellularPotentialNodes |
| unsigned | mExtracelluarColumnId |
| unsigned | mRowForAverageOfPhiZeroed |
| bool | mHasBath |
| Electrodes< DIM > * | mpElectrodes |
| BidomainDg0Assembler< DIM, DIM > * | mpAssembler |
Friends | |
| class | TestBidomainWithBathAssembler |
Detailed Description
template<unsigned DIM>
class BidomainProblem< DIM >
Class which specifies and solves a bidomain problem.The solution vector is of the form: (V_1, phi_1, V_2, phi_2, ......, V_N, phi_N), where V_j is the voltage at node j and phi_j is the extracellular potential at node j.
Definition at line 53 of file BidomainProblem.hpp.
Constructor & Destructor Documentation
| BidomainProblem< DIM >::BidomainProblem | ( | AbstractCardiacCellFactory< DIM > * | pCellFactory, | |
| bool | hasBath = false | |||
| ) | [inline] |
Constructor
- Parameters:
-
pCellFactory User defined cell factory which shows how the pde should create cells. hasBath Whether the simulation has a bath (if this is true, all elements with attribute = 1 will be set to be bath elements (the rest should have attribute = 0)).
Definition at line 177 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mFixedExtracellularPotentialNodes.
Member Function Documentation
| Vec BidomainProblem< DIM >::CreateInitialCondition | ( | ) | [inline, protected, virtual] |
Create normal initial condition but overwrite V to zero for bath nodes, if there are any.
Reimplemented from AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 87 of file BidomainProblem.cpp.
References HeartRegionCode::BATH, DistributedVector::Begin(), DistributedVectorFactory::CreateDistributedVector(), AbstractCardiacProblem< ELEM_DIM, SPACE_DIM, PROBLEM_DIM >::CreateInitialCondition(), DistributedVector::End(), AbstractMesh< ELEMENT_DIM, SPACE_DIM >::GetDistributedVectorFactory(), AbstractMesh< ELEMENT_DIM, SPACE_DIM >::GetNode(), BidomainProblem< DIM >::mHasBath, AbstractCardiacProblem< DIM, DIM, 2 >::mpMesh, and DistributedVector::Restore().
| void BidomainProblem< DIM >::AnalyseMeshForBath | ( | ) | [inline, protected] |
Annotate bath nodes with the correct region code, if a bath is present. Will throw if mHasBath is set but no bath is present in the mesh.
Definition at line 41 of file BidomainProblem.cpp.
References HeartRegionCode::BATH, AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM >::GetElementIteratorBegin(), AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM >::GetElementIteratorEnd(), AbstractElement< ELEMENT_DIM, SPACE_DIM >::GetNode(), AbstractMesh< ELEMENT_DIM, SPACE_DIM >::GetNodeIteratorBegin(), AbstractMesh< ELEMENT_DIM, SPACE_DIM >::GetNodeIteratorEnd(), AbstractElement< ELEMENT_DIM, SPACE_DIM >::GetNumNodes(), AbstractElement< ELEMENT_DIM, SPACE_DIM >::GetRegion(), BidomainProblem< DIM >::mHasBath, AbstractCardiacProblem< DIM, DIM, 2 >::mpMesh, and HeartRegionCode::TISSUE.
Referenced by BidomainProblem< DIM >::CreateCardiacPde().
| AbstractCardiacPde< DIM > * BidomainProblem< DIM >::CreateCardiacPde | ( | ) | [inline, protected, virtual] |
Create our bidomain PDE object
Implements AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 112 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::AnalyseMeshForBath(), BidomainProblem< DIM >::mpBidomainPde, and AbstractCardiacProblem< DIM, DIM, 2 >::mpCellFactory.
| AbstractDynamicAssemblerMixin< DIM, DIM, 2 > * BidomainProblem< DIM >::CreateAssembler | ( | ) | [inline, protected, virtual] |
Create a suitable bidomain assembler
Implements AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 120 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mFixedExtracellularPotentialNodes, BidomainProblem< DIM >::mHasBath, BidomainProblem< DIM >::mpAssembler, BidomainProblem< DIM >::mpBidomainPde, AbstractCardiacProblem< DIM, DIM, 2 >::mpBoundaryConditionsContainer, AbstractCardiacProblem< DIM, DIM, 2 >::mpMesh, BidomainProblem< DIM >::mRowForAverageOfPhiZeroed, AbstractCardiacProblem< DIM, DIM, 2 >::mUseMatrixBasedRhsAssembly, BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::SetFixedExtracellularPotentialNodes(), and BidomainDg0Assembler< ELEMENT_DIM, SPACE_DIM >::SetRowForAverageOfPhiZeroed().
| void BidomainProblem< DIM >::SetFixedExtracellularPotentialNodes | ( | std::vector< unsigned > | nodes | ) | [inline] |
Set the nodes at which phi_e (the extracellular potential) is fixed to zero. This does not necessarily have to be called. If it is not, phi_e is only defined up to a constant.
- Parameters:
-
nodes the nodes to be fixed.
- Note:
- currently, the value of phi_e at the fixed nodes cannot be set to be anything other than zero.
Definition at line 189 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mFixedExtracellularPotentialNodes.
| void BidomainProblem< DIM >::SetNodeForAverageOfPhiZeroed | ( | unsigned | node | ) | [inline] |
Set which row of the linear system should be used to enforce the condition that the average of phi_e is zero. If not called, this condition will not be used.
- Parameters:
-
node the mesh node index giving the row at which to impose the constraint
Definition at line 201 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mRowForAverageOfPhiZeroed.
| BidomainPde< DIM > * BidomainProblem< DIM >::GetBidomainPde | ( | ) | [inline] |
Get the pde. Can only be called after Initialise()
Definition at line 207 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mpBidomainPde.
| void BidomainProblem< DIM >::WriteInfo | ( | double | time | ) | [inline, virtual] |
Print out time and max/min voltage/phi_e values at current time.
- Parameters:
-
time current time.
Implements AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 214 of file BidomainProblem.cpp.
References AbstractMesh< ELEMENT_DIM, SPACE_DIM >::GetNumNodes(), AbstractCardiacProblem< DIM, DIM, 2 >::mpMesh, AbstractCardiacProblem< DIM, DIM, 2 >::mSolution, and ReplicatableVector::ReplicatePetscVector().
| void BidomainProblem< DIM >::DefineWriterColumns | ( | ) | [inline, virtual] |
Define what variables are written to the primary results file. Adds the extracellular potential.
Reimplemented from AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 255 of file BidomainProblem.cpp.
References Hdf5DataWriter::DefineVariable(), AbstractCardiacProblem< ELEM_DIM, SPACE_DIM, PROBLEM_DIM >::DefineWriterColumns(), BidomainProblem< DIM >::mExtracelluarColumnId, and AbstractCardiacProblem< DIM, DIM, 2 >::mpWriter.
| void BidomainProblem< DIM >::WriteOneStep | ( | double | time, | |
| Vec | voltageVec | |||
| ) | [inline, virtual] |
Write one timestep of output data to the primary results file. Adds the extracellular potential to the results.
- Parameters:
-
time the current time voltageVec the solution vector to write
Reimplemented from AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 262 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mExtracelluarColumnId, AbstractCardiacProblem< DIM, DIM, 2 >::mpWriter, AbstractCardiacProblem< DIM, DIM, 2 >::mVoltageColumnId, Hdf5DataWriter::PutStripedVector(), and Hdf5DataWriter::PutUnlimitedVariable().
| void BidomainProblem< DIM >::PreSolveChecks | ( | ) | [inline, virtual] |
Performs a series of checks before solving. Checks that a suitable method of resolving the singularity is being used.
Reimplemented from AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 269 of file BidomainProblem.cpp.
References HeartConfig::GetUseRelativeTolerance(), HeartConfig::Instance(), BidomainProblem< DIM >::mFixedExtracellularPotentialNodes, BidomainProblem< DIM >::mRowForAverageOfPhiZeroed, and AbstractCardiacProblem< ELEM_DIM, SPACE_DIM, PROBLEM_DIM >::PreSolveChecks().
| void BidomainProblem< DIM >::SetElectrodes | ( | Electrodes< DIM > & | rElectrodes | ) | [inline] |
Set an electrode object (which provides boundary conditions). Only valid if there is a bath.
- Parameters:
-
rElectrodes
Definition at line 289 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mHasBath, BidomainProblem< DIM >::mpElectrodes, and AbstractCardiacProblem< DIM, DIM, 2 >::SetBoundaryConditionsContainer().
| void BidomainProblem< DIM >::OnEndOfTimestep | ( | double | time | ) | [inline, virtual] |
Called at end of each time step in the main time-loop in AbstractCardiacProblem::Solve(). Overloaded here to switch off the electrodes (if there are any).
- Parameters:
-
time the current time
Reimplemented from AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 303 of file BidomainProblem.cpp.
References BidomainProblem< DIM >::mpAssembler, AbstractCardiacProblem< DIM, DIM, 2 >::mpBoundaryConditionsContainer, AbstractCardiacProblem< DIM, DIM, 2 >::mpDefaultBoundaryConditionsContainer, BidomainProblem< DIM >::mpElectrodes, AbstractCardiacProblem< DIM, DIM, 2 >::mpMesh, and AbstractAssembler< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetBoundaryConditionsContainer().
Member Data Documentation
BidomainPde<DIM>* BidomainProblem< DIM >::mpBidomainPde [protected] |
The bidomain PDE
Definition at line 60 of file BidomainProblem.hpp.
Referenced by BidomainProblem< DIM >::CreateAssembler(), BidomainProblem< DIM >::CreateCardiacPde(), and BidomainProblem< DIM >::GetBidomainPde().
std::vector<unsigned> BidomainProblem< DIM >::mFixedExtracellularPotentialNodes [protected] |
Nodes at which the extracellular voltage is fixed to zero (replicated)
Definition at line 63 of file BidomainProblem.hpp.
Referenced by BidomainProblem< DIM >::BidomainProblem(), BidomainProblem< DIM >::CreateAssembler(), BidomainProblem< DIM >::PreSolveChecks(), and BidomainProblem< DIM >::SetFixedExtracellularPotentialNodes().
unsigned BidomainProblem< DIM >::mExtracelluarColumnId [protected] |
Used by the writer
Definition at line 65 of file BidomainProblem.hpp.
Referenced by BidomainProblem< DIM >::DefineWriterColumns(), and BidomainProblem< DIM >::WriteOneStep().
unsigned BidomainProblem< DIM >::mRowForAverageOfPhiZeroed [protected] |
Another method of resolving the singularity in the bidomain equations. Specifies a row of the matrix at which to impose an extra condition.
Definition at line 70 of file BidomainProblem.hpp.
Referenced by BidomainProblem< DIM >::CreateAssembler(), BidomainProblem< DIM >::PreSolveChecks(), and BidomainProblem< DIM >::SetNodeForAverageOfPhiZeroed().
bool BidomainProblem< DIM >::mHasBath [protected] |
Whether the mesh has a bath, ie whether this is a bath simulation
Definition at line 73 of file BidomainProblem.hpp.
Referenced by BidomainProblem< DIM >::AnalyseMeshForBath(), BidomainProblem< DIM >::CreateAssembler(), BidomainProblem< DIM >::CreateInitialCondition(), and BidomainProblem< DIM >::SetElectrodes().
Electrodes<DIM>* BidomainProblem< DIM >::mpElectrodes [protected] |
Electrodes used to provide a shock
Definition at line 76 of file BidomainProblem.hpp.
Referenced by BidomainProblem< DIM >::OnEndOfTimestep(), and BidomainProblem< DIM >::SetElectrodes().
BidomainDg0Assembler<DIM,DIM>* BidomainProblem< DIM >::mpAssembler [protected] |
We need to save the assembler that is being used to switch off the electrodes (by adding default boundary conditions to the assembler)
Reimplemented from AbstractCardiacProblem< DIM, DIM, 2 >.
Definition at line 94 of file BidomainProblem.hpp.
Referenced by BidomainProblem< DIM >::CreateAssembler(), and BidomainProblem< DIM >::OnEndOfTimestep().
The documentation for this class was generated from the following files:
- /tmp/release_1.1/heart/src/problem/BidomainProblem.hpp
- /tmp/release_1.1/heart/src/problem/BidomainProblem.cpp
 1.5.5
1.5.5