StochasticWntCellCycleModel Class Reference
#include <StochasticWntCellCycleModel.hpp>
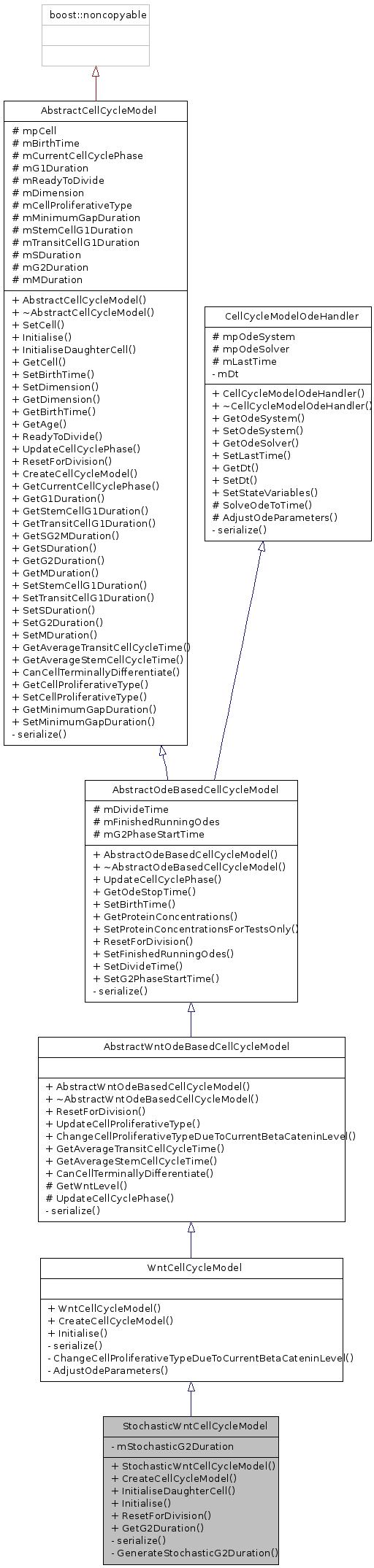
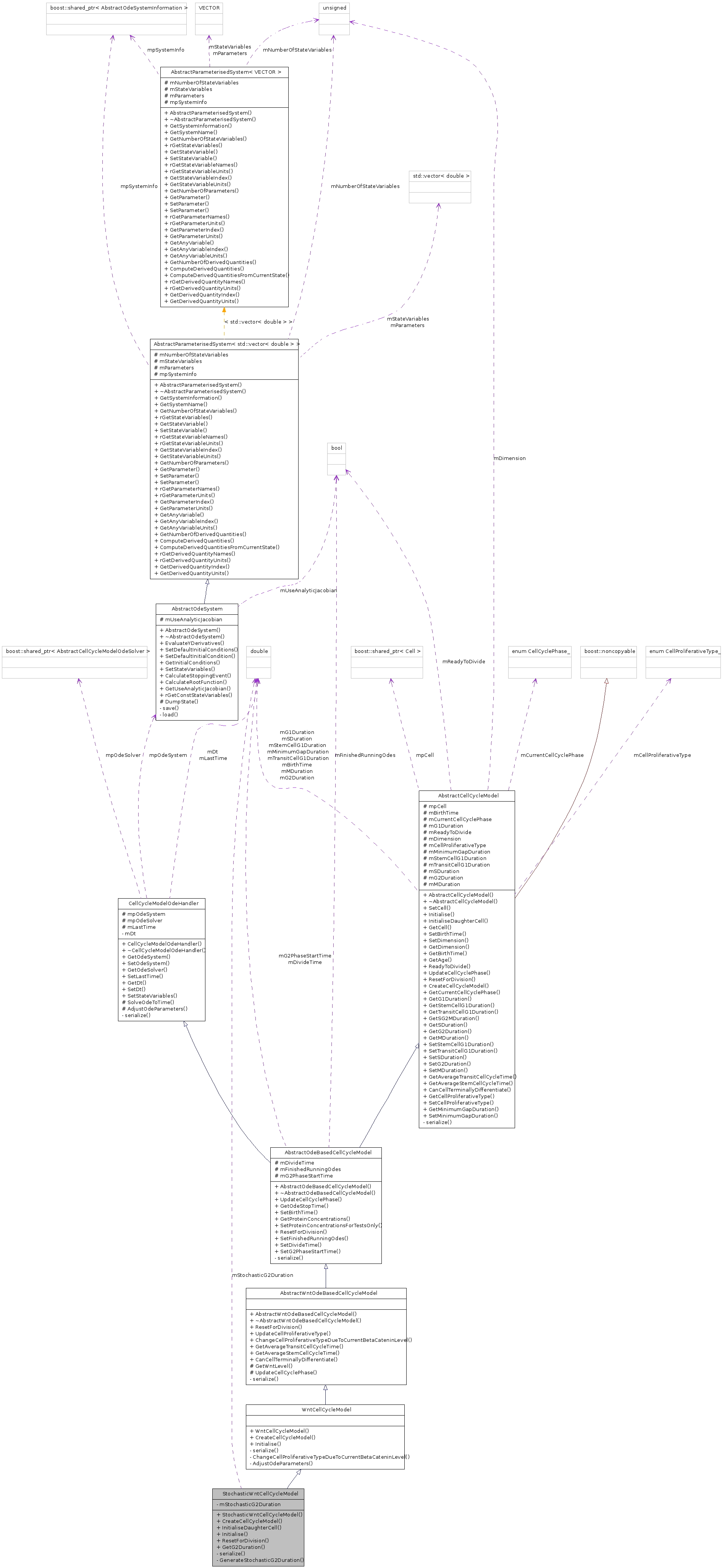
Public Member Functions | |
| StochasticWntCellCycleModel (boost::shared_ptr< AbstractCellCycleModelOdeSolver > pOdeSolver=boost::shared_ptr< AbstractCellCycleModelOdeSolver >()) | |
| AbstractCellCycleModel * | CreateCellCycleModel () |
| void | InitialiseDaughterCell () |
| void | Initialise () |
| void | ResetForDivision () |
| double | GetG2Duration () |
Private Member Functions | |
| template<class Archive> | |
| void | serialize (Archive &archive, const unsigned int version) |
| void | GenerateStochasticG2Duration () |
Private Attributes | |
| double | mStochasticG2Duration |
Friends | |
| class | boost::serialization::access |
Detailed Description
Wnt-dependent cell cycle model with a stochastic G2 duration.Note that this class uses C++'s default copying semantics, and so doesn't implement a copy constructor or operator=.
Definition at line 43 of file StochasticWntCellCycleModel.hpp.
Constructor & Destructor Documentation
| StochasticWntCellCycleModel::StochasticWntCellCycleModel | ( | boost::shared_ptr< AbstractCellCycleModelOdeSolver > | pOdeSolver = boost::shared_ptr<AbstractCellCycleModelOdeSolver>() |
) |
The standard constructor called in tests.
- Parameters:
-
pOdeSolver An optional pointer to a cell cycle model ODE solver object (allows the use of different ODE solvers)
Definition at line 31 of file StochasticWntCellCycleModel.cpp.
References CellCycleModelOdeSolver< CELL_CYCLE_MODEL, ODE_SOLVER >::Instance(), and CellCycleModelOdeHandler::mpOdeSolver.
Referenced by CreateCellCycleModel().
Member Function Documentation
| void StochasticWntCellCycleModel::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Archive the cell cycle model and member variables. Used by boost, never directly by chaste code.
Serialization of singleton objects must be done with care. Before the object is serialized via a pointer, it *MUST* be serialized directly, or an assertion will trip when a second instance of the class is created on de-serialization.
- Parameters:
-
archive the archive version the current version of this class
Reimplemented from WntCellCycleModel.
Definition at line 62 of file StochasticWntCellCycleModel.hpp.
References RandomNumberGenerator::Instance(), and mStochasticG2Duration.
| void StochasticWntCellCycleModel::GenerateStochasticG2Duration | ( | ) | [private] |
This method introduces the stochastic element of this class.
We allow the duration of the G2 phase of the cell cycle to vary as a normal random deviate with a mean of its deterministic duration, a standard deviation of 0.9 hours, and a cutoff to ensure that it is greater than some minimum value.
Definition at line 49 of file StochasticWntCellCycleModel.cpp.
References AbstractCellCycleModel::GetG2Duration(), RandomNumberGenerator::Instance(), AbstractCellCycleModel::mMinimumGapDuration, mStochasticG2Duration, and RandomNumberGenerator::NormalRandomDeviate().
Referenced by Initialise(), InitialiseDaughterCell(), and ResetForDivision().
| AbstractCellCycleModel * StochasticWntCellCycleModel::CreateCellCycleModel | ( | ) | [virtual] |
Overridden builder method to create new copies of this cell cycle model.
Reimplemented from WntCellCycleModel.
Definition at line 88 of file StochasticWntCellCycleModel.cpp.
References AbstractWntOdeBasedCellCycleModel::GetWntLevel(), AbstractCellCycleModel::mBirthTime, AbstractCellCycleModel::mCellProliferativeType, AbstractCellCycleModel::mDimension, AbstractOdeBasedCellCycleModel::mDivideTime, AbstractOdeBasedCellCycleModel::mFinishedRunningOdes, AbstractOdeBasedCellCycleModel::mG2PhaseStartTime, CellCycleModelOdeHandler::mLastTime, AbstractCellCycleModel::mpCell, CellCycleModelOdeHandler::mpOdeSolver, CellCycleModelOdeHandler::mpOdeSystem, AbstractParameterisedSystem< VECTOR >::rGetStateVariables(), AbstractOdeBasedCellCycleModel::SetBirthTime(), AbstractCellCycleModel::SetCellProliferativeType(), AbstractCellCycleModel::SetDimension(), AbstractOdeBasedCellCycleModel::SetDivideTime(), AbstractOdeBasedCellCycleModel::SetFinishedRunningOdes(), AbstractOdeBasedCellCycleModel::SetG2PhaseStartTime(), CellCycleModelOdeHandler::SetLastTime(), CellCycleModelOdeHandler::SetOdeSystem(), CellCycleModelOdeHandler::SetStateVariables(), and StochasticWntCellCycleModel().
| void StochasticWntCellCycleModel::InitialiseDaughterCell | ( | ) | [virtual] |
Set the duration of the G2 phase for the daughter cell.
Reimplemented from AbstractCellCycleModel.
Definition at line 66 of file StochasticWntCellCycleModel.cpp.
References GenerateStochasticG2Duration().
| void StochasticWntCellCycleModel::Initialise | ( | void | ) | [virtual] |
Initialise the cell cycle model at the start of a simulation.
This overridden method sets up a new WntCellCycleOdeSystem, sets the cell type according to the current beta catenin level and sets a random G2 duration.
Reimplemented from WntCellCycleModel.
Definition at line 71 of file StochasticWntCellCycleModel.cpp.
References GenerateStochasticG2Duration(), and WntCellCycleModel::Initialise().
| void StochasticWntCellCycleModel::ResetForDivision | ( | ) | [virtual] |
Reset cell cycle model by calling AbstractOdeBasedCellCycleModel::ResetForDivision() and setting a new random G2 duration.
Reimplemented from AbstractWntOdeBasedCellCycleModel.
Definition at line 77 of file StochasticWntCellCycleModel.cpp.
References GenerateStochasticG2Duration(), and AbstractWntOdeBasedCellCycleModel::ResetForDivision().
| double StochasticWntCellCycleModel::GetG2Duration | ( | ) | [virtual] |
Get the duration of the G2 phase.
Reimplemented from AbstractCellCycleModel.
Definition at line 83 of file StochasticWntCellCycleModel.cpp.
References mStochasticG2Duration.
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented from WntCellCycleModel.
Definition at line 48 of file StochasticWntCellCycleModel.hpp.
Member Data Documentation
double StochasticWntCellCycleModel::mStochasticG2Duration [private] |
The duration of the G2 phase, set stochastically.
Definition at line 74 of file StochasticWntCellCycleModel.hpp.
Referenced by GenerateStochasticG2Duration(), GetG2Duration(), and serialize().
The documentation for this class was generated from the following files:
- crypt/src/cell/cycle/StochasticWntCellCycleModel.hpp
- crypt/src/cell/cycle/StochasticWntCellCycleModel.cpp
 1.5.5
1.5.5