PropagationPropertiesCalculator Class Reference
#include <PropagationPropertiesCalculator.hpp>
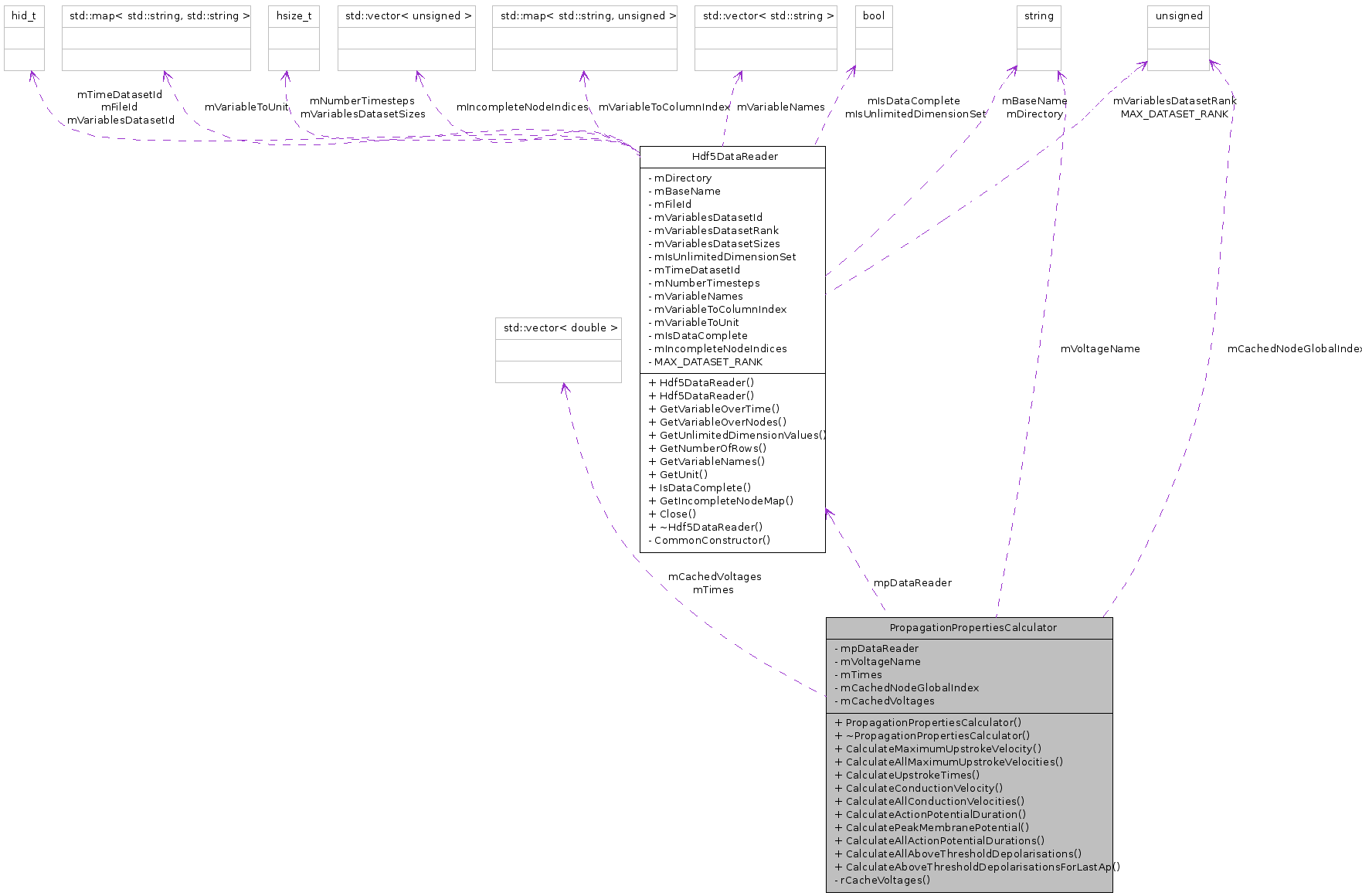
Public Member Functions | |
| PropagationPropertiesCalculator (Hdf5DataReader *pDataReader, const std::string voltageName="V") | |
| double | CalculateMaximumUpstrokeVelocity (unsigned globalNodeIndex) |
| std::vector< double > | CalculateAllMaximumUpstrokeVelocities (unsigned globalNodeIndex, double threshold) |
| std::vector< double > | CalculateUpstrokeTimes (unsigned globalNodeIndex, double threshold) |
| double | CalculateConductionVelocity (unsigned globalNearNodeIndex, unsigned globalFarNodeIndex, const double euclideanDistance) |
| std::vector< double > | CalculateAllConductionVelocities (unsigned globalNearNodeIndex, unsigned globalFarNodeIndex, const double euclideanDistance) |
| double | CalculateActionPotentialDuration (const double percentage, unsigned globalNodeIndex) |
| double | CalculatePeakMembranePotential (unsigned globalNodeIndex) |
| std::vector< double > | CalculateAllActionPotentialDurations (const double percentage, unsigned globalNodeIndex, double threshold) |
| std::vector< unsigned > | CalculateAllAboveThresholdDepolarisations (unsigned globalNodeIndex, double threshold) |
| unsigned | CalculateAboveThresholdDepolarisationsForLastAp (unsigned globalNodeIndex, double threshold) |
Private Member Functions | |
| std::vector< double > & | rCacheVoltages (unsigned globalNodeIndex) |
Private Attributes | |
| Hdf5DataReader * | mpDataReader |
| const std::string | mVoltageName |
| std::vector< double > | mTimes |
| unsigned | mCachedNodeGlobalIndex |
| std::vector< double > | mCachedVoltages |
Detailed Description
Calculate physiological properties at given global mesh indices- maximum upstroke velocity at a single cell
- times of upstroke at a single cell
- (all) conduction velocities between two cells
- (all) action potential duration at a single cell
- maximum transmembrane potential (maximum systolic potential) at a single cell.
Definition at line 44 of file PropagationPropertiesCalculator.hpp.
Constructor & Destructor Documentation
| PropagationPropertiesCalculator::PropagationPropertiesCalculator | ( | Hdf5DataReader * | pDataReader, | |
| const std::string | voltageName = "V" | |||
| ) |
Constructor.
- Parameters:
-
pDataReader Pointer to the data reader containing the simulation. voltageName Optionally the name of the variable representing the membrane potential. Defaults to "V".
Definition at line 35 of file PropagationPropertiesCalculator.cpp.
Member Function Documentation
| std::vector< double > & PropagationPropertiesCalculator::rCacheVoltages | ( | unsigned | globalNodeIndex | ) | [private] |
Read the voltages vector for the given node and cache it, returning a reference to the cached vector. If subsequently called with the same index, will return the cached vector without re-reading from file.
Note: will only cache the last node index used.
- Parameters:
-
globalNodeIndex the index of the node to cache voltages for
Definition at line 239 of file PropagationPropertiesCalculator.cpp.
References Hdf5DataReader::GetVariableOverTime(), mCachedNodeGlobalIndex, mCachedVoltages, mpDataReader, and mVoltageName.
Referenced by CalculateAboveThresholdDepolarisationsForLastAp(), CalculateActionPotentialDuration(), CalculateAllAboveThresholdDepolarisations(), CalculateAllActionPotentialDurations(), CalculateAllConductionVelocities(), CalculateAllMaximumUpstrokeVelocities(), CalculateConductionVelocity(), CalculateMaximumUpstrokeVelocity(), CalculatePeakMembranePotential(), and CalculateUpstrokeTimes().
| double PropagationPropertiesCalculator::CalculateMaximumUpstrokeVelocity | ( | unsigned | globalNodeIndex | ) |
Calculate the maximum upstroke velocity at a single cell. We calculate for the last upstroke found in the simulation data.
- Parameters:
-
globalNodeIndex The cell at which to calculate.
Definition at line 48 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetLastMaxUpstrokeVelocity(), mTimes, and rCacheVoltages().
| std::vector< double > PropagationPropertiesCalculator::CalculateAllMaximumUpstrokeVelocities | ( | unsigned | globalNodeIndex, | |
| double | threshold | |||
| ) |
Calculate the maximum upstroke velocity at a single cell. We return all the max upstroke velocities for all APs.
- Parameters:
-
globalNodeIndex The cell at which to calculate. threshold The voltage threshold (we use this for marking the end of an AP)
Definition at line 55 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetMaxUpstrokeVelocities(), mTimes, and rCacheVoltages().
Referenced by PostProcessingWriter< ELEMENT_DIM, SPACE_DIM >::WriteAboveThresholdDepolarisationFile(), and PostProcessingWriter< ELEMENT_DIM, SPACE_DIM >::WriteMaxUpstrokeVelocityMap().
| std::vector< double > PropagationPropertiesCalculator::CalculateUpstrokeTimes | ( | unsigned | globalNodeIndex, | |
| double | threshold | |||
| ) |
Calculate the times of upstroke at a single cell. We return all the times of upstroke velocities for all APs.
- Parameters:
-
globalNodeIndex The cell at which to calculate. threshold The voltage threshold for APD calculation (we count this as the start of an AP)
Definition at line 62 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetTimesAtMaxUpstrokeVelocity(), mTimes, and rCacheVoltages().
Referenced by PostProcessingWriter< ELEMENT_DIM, SPACE_DIM >::WriteUpstrokeTimeMap().
| double PropagationPropertiesCalculator::CalculateConductionVelocity | ( | unsigned | globalNearNodeIndex, | |
| unsigned | globalFarNodeIndex, | |||
| const double | euclideanDistance | |||
| ) |
Calculate the conduction velocity between two cells, i.e. the time taken for an AP to propagate from one to the other. It returns the value of conduction velocity of the LAST action potential that reached both nodes. Throws exceptions if an AP never reached one of the nodes.
- Parameters:
-
globalNearNodeIndex The cell to measure from. globalFarNodeIndex The cell to measure to. euclideanDistance The distance the AP travels between the cells, along the tissue.
Definition at line 104 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetMaxUpstrokeVelocities(), CellProperties::GetTimeAtLastMaxUpstrokeVelocity(), CellProperties::GetTimesAtMaxUpstrokeVelocity(), Hdf5DataReader::GetVariableOverTime(), mpDataReader, mTimes, mVoltageName, and rCacheVoltages().
Referenced by AbstractConvergenceTester< CELL, CARDIAC_PROBLEM, DIM, PROBLEM_DIM >::Converge().
| std::vector< double > PropagationPropertiesCalculator::CalculateAllConductionVelocities | ( | unsigned | globalNearNodeIndex, | |
| unsigned | globalFarNodeIndex, | |||
| const double | euclideanDistance | |||
| ) |
Calculate all the conduction velocities between two cells, i.e. the time taken for all APs to propagate from one to the other. It returns a vector containing all the conduction velocities for each of the APs that reached the two nodes (only the APs that reached both nodes). Throws exceptions if an AP never reached one of the nodes.
- Parameters:
-
globalNearNodeIndex The cell to measure from. globalFarNodeIndex The cell to measure to. euclideanDistance The distance the AP travels between the cells, along the tissue.
Definition at line 159 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetTimesAtMaxUpstrokeVelocity(), Hdf5DataReader::GetVariableOverTime(), mpDataReader, mTimes, mVoltageName, and rCacheVoltages().
Referenced by PostProcessingWriter< ELEMENT_DIM, SPACE_DIM >::WriteConductionVelocityMap().
| double PropagationPropertiesCalculator::CalculateActionPotentialDuration | ( | const double | percentage, | |
| unsigned | globalNodeIndex | |||
| ) |
Calculate the action potential duration at a single cell. We calculate for the last AP found in the simulation data.
- Parameters:
-
percentage The percentage of the amplitude to calculate for. globalNodeIndex The cell at which to calculate.
Definition at line 69 of file PropagationPropertiesCalculator.cpp.
References EXCEPTION, CellProperties::GetLastActionPotentialDuration(), mTimes, and rCacheVoltages().
Referenced by AbstractConvergenceTester< CELL, CARDIAC_PROBLEM, DIM, PROBLEM_DIM >::Converge().
| double PropagationPropertiesCalculator::CalculatePeakMembranePotential | ( | unsigned | globalNodeIndex | ) |
Calculate the maximum transmembrane potential (maximum systolic potential) at a single cell. We calculate for the last AP found in the simulation data.
- Parameters:
-
globalNodeIndex The cell at which to calculate.
Definition at line 90 of file PropagationPropertiesCalculator.cpp.
References rCacheVoltages().
| std::vector< double > PropagationPropertiesCalculator::CalculateAllActionPotentialDurations | ( | const double | percentage, | |
| unsigned | globalNodeIndex, | |||
| double | threshold | |||
| ) |
Calculate all the action potentials duration at a single cell.
- Parameters:
-
percentage The percentage of the amplitude to calculate for. globalNodeIndex The cell at which to calculate. threshold The voltage threshold for APD calculation (we count this as the start of an AP)
Definition at line 81 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetAllActionPotentialDurations(), mTimes, and rCacheVoltages().
Referenced by PostProcessingWriter< ELEMENT_DIM, SPACE_DIM >::WriteApdMapFile().
| std::vector< unsigned > PropagationPropertiesCalculator::CalculateAllAboveThresholdDepolarisations | ( | unsigned | globalNodeIndex, | |
| double | threshold | |||
| ) |
Calculates all the depolarisations that occur above threshold at a single cell.
- Parameters:
-
globalNodeIndex the cell at which to calculate threshold the threshold above which the depolarisations are counted
Definition at line 223 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetNumberOfAboveThresholdDepolarisationsForAllAps(), mTimes, and rCacheVoltages().
Referenced by PostProcessingWriter< ELEMENT_DIM, SPACE_DIM >::WriteAboveThresholdDepolarisationFile().
| unsigned PropagationPropertiesCalculator::CalculateAboveThresholdDepolarisationsForLastAp | ( | unsigned | globalNodeIndex, | |
| double | threshold | |||
| ) |
Calculates the depolarisations that occur above threshold at a single cell during the last recorded Ap
- Parameters:
-
globalNodeIndex the cell at which to calculate threshold the threshold above which the depolarisations are counted
Definition at line 231 of file PropagationPropertiesCalculator.cpp.
References CellProperties::GetNumberOfAboveThresholdDepolarisationsForLastAp(), mTimes, and rCacheVoltages().
Member Data Documentation
Reader to get the data from which we use to calculate properties.
Definition at line 48 of file PropagationPropertiesCalculator.hpp.
Referenced by CalculateAllConductionVelocities(), CalculateConductionVelocity(), and rCacheVoltages().
const std::string PropagationPropertiesCalculator::mVoltageName [private] |
Name of the variable representing the membrane potential.
Definition at line 50 of file PropagationPropertiesCalculator.hpp.
Referenced by CalculateAllConductionVelocities(), CalculateConductionVelocity(), and rCacheVoltages().
std::vector<double> PropagationPropertiesCalculator::mTimes [private] |
Time values
Definition at line 52 of file PropagationPropertiesCalculator.hpp.
Referenced by CalculateAboveThresholdDepolarisationsForLastAp(), CalculateActionPotentialDuration(), CalculateAllAboveThresholdDepolarisations(), CalculateAllActionPotentialDurations(), CalculateAllConductionVelocities(), CalculateAllMaximumUpstrokeVelocities(), CalculateConductionVelocity(), CalculateMaximumUpstrokeVelocity(), and CalculateUpstrokeTimes().
unsigned PropagationPropertiesCalculator::mCachedNodeGlobalIndex [private] |
Which node voltages have been cached for, if any
Definition at line 54 of file PropagationPropertiesCalculator.hpp.
Referenced by rCacheVoltages().
std::vector<double> PropagationPropertiesCalculator::mCachedVoltages [private] |
The cached voltages vector
Definition at line 56 of file PropagationPropertiesCalculator.hpp.
Referenced by rCacheVoltages().
The documentation for this class was generated from the following files:
- heart/src/postprocessing/PropagationPropertiesCalculator.hpp
- heart/src/postprocessing/PropagationPropertiesCalculator.cpp
 1.5.5
1.5.5