ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel Class Reference
#include <ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.hpp>
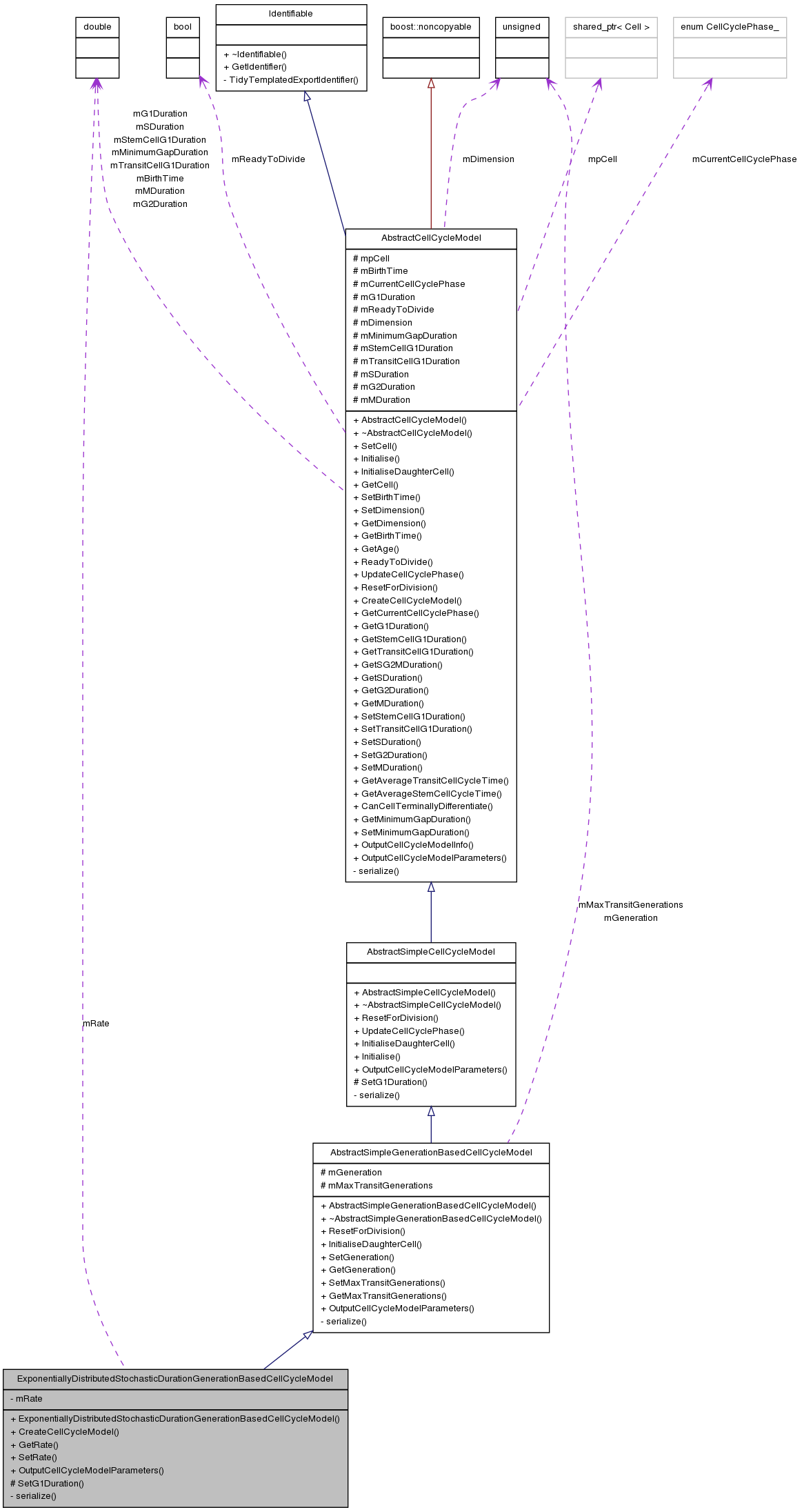
Inherits AbstractSimpleGenerationBasedCellCycleModel.
Public Member Functions | |
| ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel () | |
| AbstractCellCycleModel * | CreateCellCycleModel () |
| double | GetRate () |
| void | SetRate (double rate) |
| virtual void | OutputCellCycleModelParameters (out_stream &rParamsFile) |
Protected Member Functions | |
| void | SetG1Duration () |
Private Member Functions | |
| template<class Archive > | |
| void | serialize (Archive &archive, const unsigned int version) |
Private Attributes | |
| double | mRate |
Friends | |
| class | TestSimpleCellCycleModels |
| class | boost::serialization::access |
Detailed Description
A cell cycle model where the G1 duration is drawn from an exponential random distribution
Definition at line 45 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.hpp.
Constructor & Destructor Documentation
| ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel::ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel | ( | ) |
Constructor - just a default, mBirthTime is now set in the AbstractCellCycleModel class. mG1Duration is set very high, it is set for the individual cells when InitialiseDaughterCell is called
Definition at line 42 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.cpp.
Referenced by CreateCellCycleModel().
Member Function Documentation
| AbstractCellCycleModel * ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel::CreateCellCycleModel | ( | ) | [virtual] |
Overridden builder method to create new copies of this cell-cycle model.
- Returns:
- new cell-cycle model
Implements AbstractCellCycleModel.
Definition at line 48 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.cpp.
References ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel(), AbstractCellCycleModel::mBirthTime, AbstractCellCycleModel::mG2Duration, AbstractSimpleGenerationBasedCellCycleModel::mGeneration, AbstractSimpleGenerationBasedCellCycleModel::mMaxTransitGenerations, AbstractCellCycleModel::mMDuration, AbstractCellCycleModel::mMinimumGapDuration, mRate, AbstractCellCycleModel::mSDuration, AbstractCellCycleModel::mStemCellG1Duration, AbstractCellCycleModel::mTransitCellG1Duration, AbstractCellCycleModel::SetBirthTime(), AbstractCellCycleModel::SetG2Duration(), AbstractSimpleGenerationBasedCellCycleModel::SetGeneration(), AbstractSimpleGenerationBasedCellCycleModel::SetMaxTransitGenerations(), AbstractCellCycleModel::SetMDuration(), AbstractCellCycleModel::SetMinimumGapDuration(), SetRate(), AbstractCellCycleModel::SetSDuration(), AbstractCellCycleModel::SetStemCellG1Duration(), and AbstractCellCycleModel::SetTransitCellG1Duration().
| double ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel::GetRate | ( | ) |
Return the rate parameter (lambda) of the exponential distribution.
- Returns:
- mRate, the rate parameter of the distribution
Definition at line 104 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.cpp.
References mRate.
| void ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel::OutputCellCycleModelParameters | ( | out_stream & | rParamsFile | ) | [virtual] |
Outputs cell cycle model parameters to file.
- Parameters:
-
rParamsFile the file stream to which the parameters are output
Reimplemented from AbstractSimpleGenerationBasedCellCycleModel.
Definition at line 118 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.cpp.
References mRate.
| void ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Archive the cell-cycle model and random number generator, never used directly - boost uses this.
- Parameters:
-
archive the archive version the current version of this class
Reimplemented from AbstractSimpleGenerationBasedCellCycleModel.
Definition at line 67 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.hpp.
References SerializableSingleton< SINGLETON_CLASS >::GetSerializationWrapper(), RandomNumberGenerator::Instance(), and mRate.
| void ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel::SetG1Duration | ( | ) | [protected, virtual] |
Stochastically set the G1 duration following an exponential distribution. Called on cell creation at the start of a simulation, and for both parent and daughter cells at cell division.
Reimplemented from AbstractSimpleCellCycleModel.
Definition at line 84 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.cpp.
References RandomNumberGenerator::ExponentialRandomDeviate(), RandomNumberGenerator::Instance(), AbstractCellCycleModel::mG1Duration, AbstractCellCycleModel::mpCell, mRate, and NEVER_REACHED.
| void ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel::SetRate | ( | double | rate | ) |
Set the rate parameter of the exponential distribution. For consistency, this function also resets the internal values for mTransitCellG1Duration and mStemCellG1Duration to the average value of the exponential random variable, i.e. 1.0/rate. This ensures that GetAverageTransitCellCycleTime and GetAverageStemCellCycleTime returns correct values.
- Parameters:
-
rate
Definition at line 109 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.cpp.
References mRate, AbstractCellCycleModel::SetStemCellG1Duration(), and AbstractCellCycleModel::SetTransitCellG1Duration().
Referenced by CreateCellCycleModel().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented from AbstractSimpleGenerationBasedCellCycleModel.
Definition at line 59 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.hpp.
Member Data Documentation
The rate parameter of the exponential distribution, often denoted by lambda. This parameter is initialised to 1.0/mTransitCellG1Duration. The latter variable is inherited from the abstract cell cycle model. This initialisation ensures that cells will have the standard G1 duration (as defined in the abstract cell cycle model classes) on average.
Definition at line 56 of file ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.hpp.
Referenced by CreateCellCycleModel(), GetRate(), OutputCellCycleModelParameters(), serialize(), SetG1Duration(), and SetRate().
The documentation for this class was generated from the following files:
- cell_based/src/population/cell/cycle/ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.hpp
- cell_based/src/population/cell/cycle/ExponentiallyDistributedStochasticDurationGenerationBasedCellCycleModel.cpp
 1.6.2
1.6.2