Cell-based Chaste
Introduction and aims
Mathematical and computational models of biological and physiological systems are rapidly increasing in complexity. This is especially true of fields such as cancer modelling, where the amount of available biological data is increasing exponentially.
Modelling approaches therefore span the range from detailed models of molecular level processes, right through to biomechanical models at the tissue level. The aim of cell-based Chaste is to develop a computational framework that bridges across these spatial and temporal scales within a single, generic modelling framework.
The initial focus of this work was the dynamic mechanisms underlying the onset of colorectal cancers. This area was chosen due to the availability of particularly good experimental data, and since its biological understanding is sufficiently advanced to allow such a systems-level approach. Colorectal cancers originate from the epithelium that covers the luminal surface of the intestinal tract. This epithelium renews itself through a coordinated programme of cell proliferation, migration and differentiation, which begins in the of tiny crypts of Lieberkühn that descend from the epithelium into the underlying connective tissue.
A specialist crypt code component has been developed in Chaste to study intestinal crypts and the initiation of colorectal cancer. This component includes code to define the intestinal crypt geometry, Wnt signalling pathway and intestinal cell-cycle models, and has been used to examine theoretically the concept and role of stem cells in crypt homeostasis and the role of mechanical effects in cell behaviour.
Implementation
We employ a multi-scale modelling framework comprising up to three interlinked modules:
- a model of cellular behaviour (for example, progress through the cell cycle, or programmed cell death);
- a model of the movement and mechanical interaction between cells; and
- a model of the transport of key nutrients, signalling molecules or waste products.
Models of sub-cellular behaviour may be either deterministic or stochastic, and range from simple rule-based models to highly non-linear ordinary differential equation models. A variety of discrete model frameworks can be used to describe the movement and mechanical interactions of cell aggregates. Chaste currently supports the simulation of cell populations using lattice-based approaches (cellular automata and the cellular Potts model) as well as lattice-free models (centre-based models, in which connectivity is defined either by a Voronoi tessellation or ‘overlapping spheres’ approach, and vertex-based models). The transport of nutrients, signalling molecules or waste products may be modelled using a continuum approach; due to the presence of cell growth and division, such partial differential equation models must be solved on an irregular and evolving domain.
User functionality
- Simulation of both cell-centre- and vertex-based models of cell populations in one, two or three spatial dimensions.
- Ability to define different force laws for cell-cell interactions.
- Ability to define different boundary conditions and geometric domains.
- Ability to define different cell killer objects, which govern the conditions in which cells undergo necrosis/apoptosis.
- Ability to define different cell-cycle models, which govern cell proliferation and quiescence.
- Ability to define different cell proliferative types (stem, transit amplifying and differentiated cells).
- Ability to define different cell mutation states, which can effect cell proliferation, migration and adhesion.
- Ability to define reaction-diffusion equations for key nutrients or signalling molecules, whose concentrations may influence (and in turn be influenced by) any of the above.
- Visualization and post-processing of simulation results.
Example simulation results
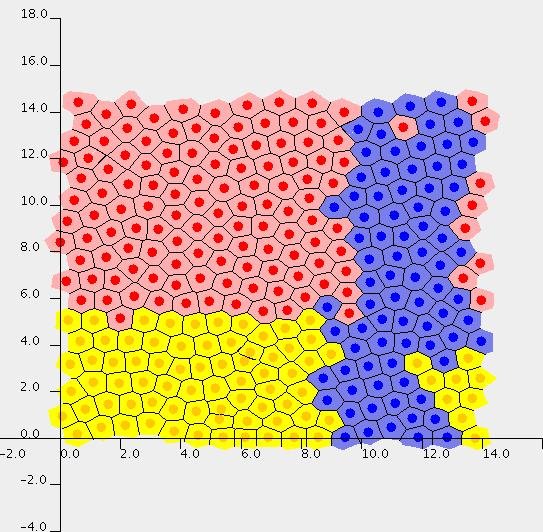
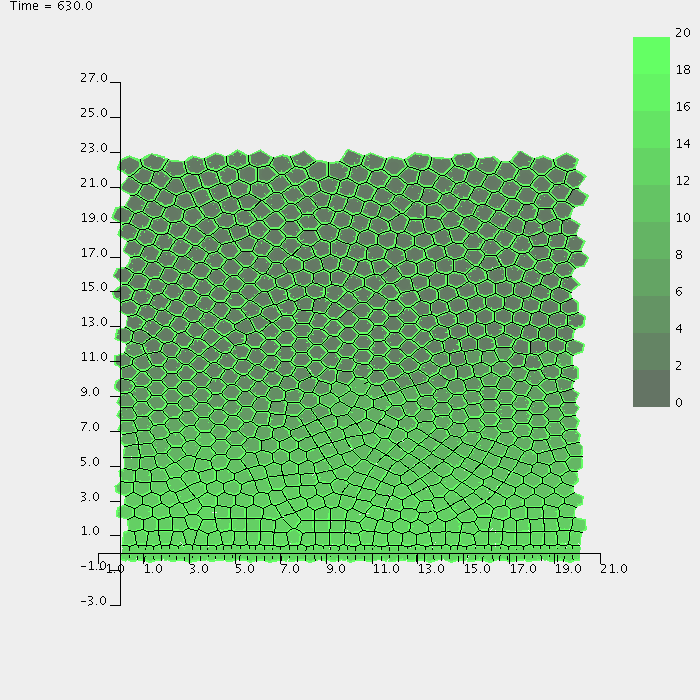
Screenshots

Videos
In addition to the examples shown below, you can find a variety of movies of simulations using the cancer and cell-based Chaste code on the Chaste YouTube channel.
A crypt represented by a cell-centre model (left) and a cell-vertex model (right):
A crypt represented by a cell-centre model with a population of mutant cells shown in black, the simulation on the right represents mutant cells with increased adhesiveness.
Monoclonal conversion in a healthy crypt using a cylindrical geometry.
Cross-sectional simulation of a crypt including stromal cells and basement membrane.
Monoclonal conversion in a healthy crypt in a three-dimensional fixed geometry.
Demonstration of a simulation with multiple crypts.
