AbstractCellCycleModel Class Reference
#include <AbstractCellCycleModel.hpp>
Inherited by AbstractOdeBasedCellCycleModel, and AbstractSimpleCellCycleModel.
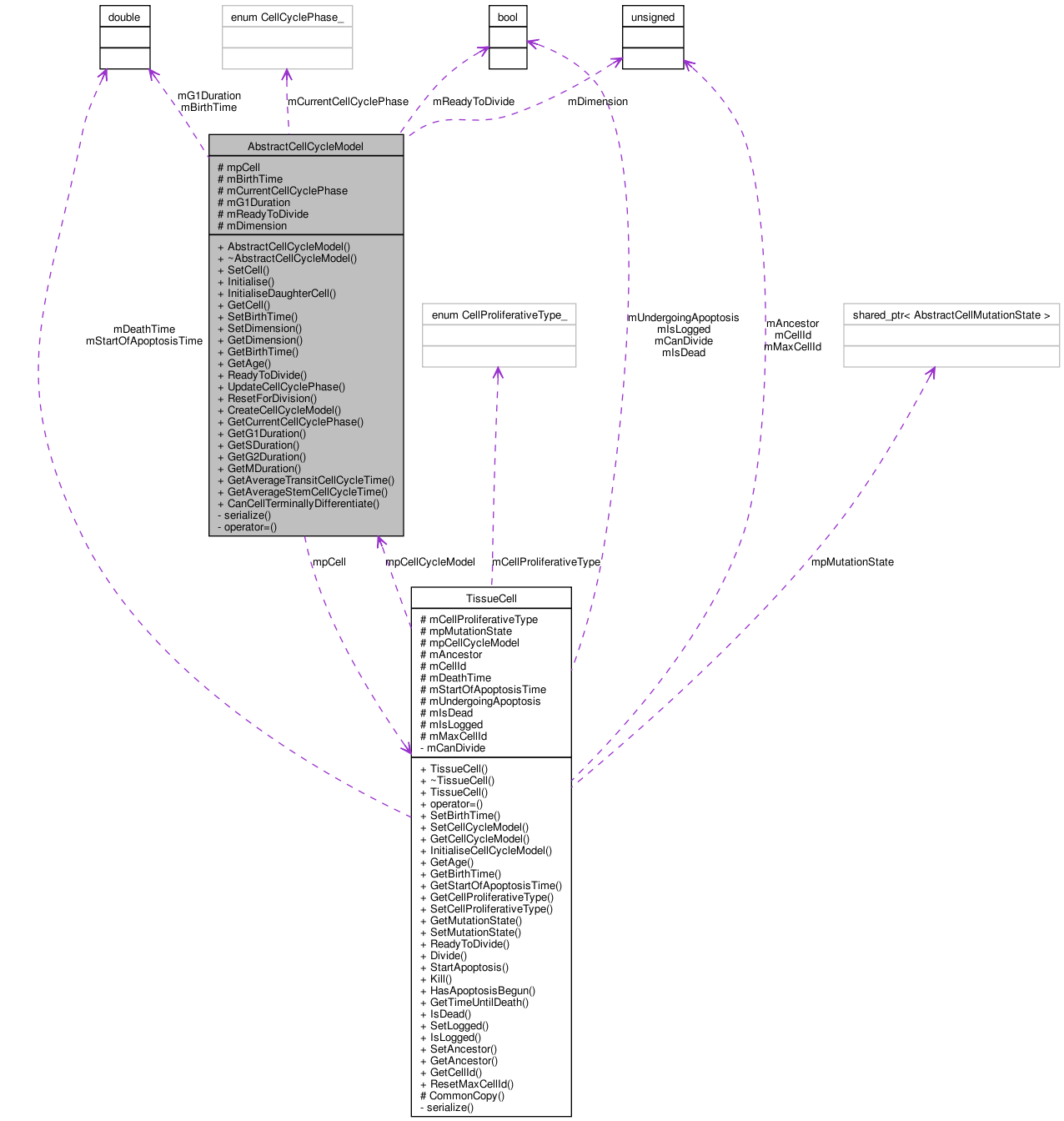
Detailed Description
The AbstractCellCycleModel contains basic information to all cell cycle models. It handles assignment of birth time, cell cycle phase and a TissueCell.
Definition at line 51 of file AbstractCellCycleModel.hpp.
Constructor & Destructor Documentation
| AbstractCellCycleModel::AbstractCellCycleModel | ( | ) |
Sets up a new AbstractCellCycleModel, gives it a birth time of the current simulation time (which is overwritten by some subclasses)
Definition at line 30 of file AbstractCellCycleModel.cpp.
| AbstractCellCycleModel::~AbstractCellCycleModel | ( | ) | [virtual] |
Base class with virtual methods needs a virtual destructor.
Definition at line 40 of file AbstractCellCycleModel.cpp.
Member Function Documentation
| bool AbstractCellCycleModel::CanCellTerminallyDifferentiate | ( | ) | [virtual] |
Whether a cell with this cell cyclde model is able to fully (terminally) differentiate.
Reimplemented in AbstractWntOdeBasedCellCycleModel, SimpleWntCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 147 of file AbstractCellCycleModel.cpp.
| virtual AbstractCellCycleModel* AbstractCellCycleModel::CreateCellCycleModel | ( | ) | [pure virtual] |
Builder method to create new instances of the cell cycle model. Each concrete subclass must implement this method to create an instance of that subclass.
This method is called by the copy constructor and operator= of TissueCell to create a copy of the cell cycle model when copying a cell. It thus needs to create an instance of the right class which is an exact copy of this instance.
This method is also called by TissueCell::Divide() to create a cell cycle model for the daughter cell. Note that the parent cell cycle model will have had ResetForDivision() called just before CreateCellCycleModel() is called, so performing an exact copy of the parent is suitable behaviour. Any daughter-cell-specific initialisation can be done in InitialiseDaughterCell().
It is suggested to implement this method using the copy constructor, for example: return new TysonNovakCellCycleModel(*this); If any special copying behaviour is required, a suitable copy constructor can then be written (which you should have done anyway, of course).
- Note:
- This base class does not define a copy constructor, despite the fact that it contains a pointer to a TissueCell. This is OK because the TissueCell is not deleted by our destructor, and in all cases where the copy constructor is used either the original object is immediately destroyed, or the copy is assigned to a new cell.
Implemented in SimpleWntCellCycleModel, SingleOdeWntCellCycleModel, StochasticWntCellCycleModel, VanLeeuwen2009WntSwatCellCycleModelHypothesisOne, VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo, WntCellCycleModel, FixedDurationGenerationBasedCellCycleModel, StochasticDurationGenerationBasedCellCycleModel, and TysonNovakCellCycleModel.
Referenced by TissueCell::CommonCopy(), and TissueCell::Divide().
| double AbstractCellCycleModel::GetAge | ( | ) |
Returns the cell's age.
Definition at line 68 of file AbstractCellCycleModel.cpp.
References SimulationTime::GetTime(), SimulationTime::Instance(), and mBirthTime.
Referenced by TissueCell::GetAge(), ReadyToDivide(), AbstractSimpleCellCycleModel::UpdateCellCyclePhase(), and AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase().
| double AbstractCellCycleModel::GetAverageStemCellCycleTime | ( | ) | [virtual] |
Return the typical cell cycle duration for a stem cell, in hours. This method is overridden in some subclasses.
Reimplemented in AbstractWntOdeBasedCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 141 of file AbstractCellCycleModel.cpp.
References TissueConfig::GetSG2MDuration(), TissueConfig::GetStemCellG1Duration(), and TissueConfig::Instance().
| double AbstractCellCycleModel::GetAverageTransitCellCycleTime | ( | ) | [virtual] |
Return the typical cell cycle duration for a transit cell, in hours. This method is overridden in some subclasses.
Reimplemented in AbstractWntOdeBasedCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 135 of file AbstractCellCycleModel.cpp.
References TissueConfig::GetSG2MDuration(), TissueConfig::GetTransitCellG1Duration(), and TissueConfig::Instance().
| double AbstractCellCycleModel::GetBirthTime | ( | ) | const |
- Returns:
- the time at which the cell was born.
Definition at line 63 of file AbstractCellCycleModel.cpp.
References mBirthTime.
Referenced by TissueCell::GetBirthTime().
| TissueCell * AbstractCellCycleModel::GetCell | ( | ) |
- Returns:
- The cell which plays host to this cell cycle model.
Definition at line 52 of file AbstractCellCycleModel.cpp.
References mpCell.
Referenced by MeshBasedTissue< DIM >::CheckTissueCellPointers().
| CellCyclePhase AbstractCellCycleModel::GetCurrentCellCyclePhase | ( | ) |
- Returns:
- the current cell cycle phase
Definition at line 73 of file AbstractCellCycleModel.cpp.
References mCurrentCellCyclePhase.
Referenced by AbstractTissue< DIM >::GenerateCellResults().
| unsigned AbstractCellCycleModel::GetDimension | ( | ) |
Get the dimension this cell cycle model thinks the simulation is in.
Definition at line 130 of file AbstractCellCycleModel.cpp.
References mDimension.
| double AbstractCellCycleModel::GetG1Duration | ( | ) | [virtual] |
- Returns:
- the duration of the G1 phase of the cell cycle
Definition at line 90 of file AbstractCellCycleModel.cpp.
References mG1Duration.
Referenced by ReadyToDivide().
| double AbstractCellCycleModel::GetG2Duration | ( | ) | [virtual] |
- Returns:
- the duration of the G2 phase of the cell cycle
Reimplemented in StochasticWntCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 95 of file AbstractCellCycleModel.cpp.
References TissueConfig::GetG2Duration(), and TissueConfig::Instance().
Referenced by ReadyToDivide(), AbstractSimpleCellCycleModel::UpdateCellCyclePhase(), and AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase().
| double AbstractCellCycleModel::GetMDuration | ( | ) | [virtual] |
- Returns:
- the duration of the M phase of the cell cycle
Reimplemented in TysonNovakCellCycleModel.
Definition at line 100 of file AbstractCellCycleModel.cpp.
References TissueConfig::GetMDuration(), and TissueConfig::Instance().
Referenced by ReadyToDivide(), AbstractSimpleCellCycleModel::UpdateCellCyclePhase(), and AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase().
| double AbstractCellCycleModel::GetSDuration | ( | ) | [virtual] |
- Returns:
- the duration of the S phase of the cell cycle
Reimplemented in TysonNovakCellCycleModel.
Definition at line 85 of file AbstractCellCycleModel.cpp.
References TissueConfig::GetSDuration(), and TissueConfig::Instance().
Referenced by ReadyToDivide(), AbstractSimpleCellCycleModel::UpdateCellCyclePhase(), and AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase().
| virtual void AbstractCellCycleModel::Initialise | ( | void | ) | [inline, virtual] |
Initialise the cell cycle model at the start of a simulation.
This method will be called precisely once per cell set up in the initial tissue. It is not called on cell division; use ResetForDivision(), CreateCellCycleModel() and InitialiseDaughterCell() for that.
By the time this is called, a Tissue will have been set up, so the model can know where its cell is located in space. If relevant to the simulation, the CellwiseData and WntConcentration singletons will also have been initialised.
Reimplemented in AbstractVanLeeuwen2009WntSwatCellCycleModel, SingleOdeWntCellCycleModel, StochasticWntCellCycleModel, WntCellCycleModel, and AbstractSimpleCellCycleModel.
Definition at line 161 of file AbstractCellCycleModel.hpp.
Referenced by TissueCell::InitialiseCellCycleModel().
| virtual void AbstractCellCycleModel::InitialiseDaughterCell | ( | ) | [inline, virtual] |
Initialise the new daughter cell's cycle model after a cell division.
This is called by TissueCell::Divide once the new cell object has been fully created, to perform any initialisation of the cell cycle which requires access to the cell.
Note that much initialisation can be performed using the combination of ResetForDivision() (called on the parent prior to division) and CreateCellCycleModel() (called on the reset parent to create the new cell cycle model object).
Reimplemented in SimpleWntCellCycleModel, StochasticWntCellCycleModel, AbstractSimpleCellCycleModel, AbstractSimpleGenerationBasedCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 176 of file AbstractCellCycleModel.hpp.
Referenced by TissueCell::Divide().
| AbstractCellCycleModel& AbstractCellCycleModel::operator= | ( | const AbstractCellCycleModel & | rOtherModel | ) | [private] |
Assignment operator has no definition and can't be called. This is to prevent running the default assignment operator.
- Parameters:
-
rOtherModel the cell cycle model being copied.
| bool AbstractCellCycleModel::ReadyToDivide | ( | ) |
Determine whether the cell is ready to divide (enter M phase).
The intention is that this method is called precisely once at each timestep of the simulation. However this does not appear to always be the case at present, and so it can cope with more unusual usage patterns.
Definition at line 105 of file AbstractCellCycleModel.cpp.
References GetAge(), GetG1Duration(), GetG2Duration(), GetMDuration(), GetSDuration(), mCurrentCellCyclePhase, mpCell, mReadyToDivide, and UpdateCellCyclePhase().
Referenced by TissueCell::ReadyToDivide().
| void AbstractCellCycleModel::ResetForDivision | ( | ) | [virtual] |
Each cell cycle model must be able to be reset 'after' a cell division.
Actually, this method is called from TissueCell::Divide() to reset the cell cycle just before the daughter cell is created. CreateCellCycleModel() can then clone our state to generate a cell cycle model instance for the daughter cell.
Reimplemented in AbstractWntOdeBasedCellCycleModel, StochasticWntCellCycleModel, AbstractOdeBasedCellCycleModel, AbstractSimpleCellCycleModel, AbstractSimpleGenerationBasedCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 78 of file AbstractCellCycleModel.cpp.
References mCurrentCellCyclePhase, and mReadyToDivide.
Referenced by TissueCell::Divide().
| void AbstractCellCycleModel::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Archive the member variables.
Serialization of singleton objects must be done with care. Before the object is serialized via a pointer, it *MUST* be serialized directly, or an assertion will trip when a second instance of the class is created on de-serialization.
- Parameters:
-
archive the archive version the current version of this class
Reimplemented in AbstractWntOdeBasedCellCycleModel, SimpleWntCellCycleModel, StochasticWntCellCycleModel, VanLeeuwen2009WntSwatCellCycleModelHypothesisOne, VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo, AbstractOdeBasedCellCycleModel, AbstractOdeBasedCellCycleModelWithStoppingEvent, AbstractSimpleCellCycleModel, AbstractSimpleGenerationBasedCellCycleModel, FixedDurationGenerationBasedCellCycleModel, StochasticDurationGenerationBasedCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 69 of file AbstractCellCycleModel.hpp.
References TissueConfig::Instance(), SimulationTime::Instance(), mBirthTime, mCurrentCellCyclePhase, mDimension, mG1Duration, and mReadyToDivide.
| void AbstractCellCycleModel::SetBirthTime | ( | double | birthTime | ) | [virtual] |
Set the cell's time of birth (usually not required as it should be inside the indivdual cell-cycle-model-constructor, but useful for tests).
- Parameters:
-
birthTime the simulation time at this cell's birth.
(This function is overridden in AbstractOdeBasedCellCycleModel).
Reimplemented in AbstractOdeBasedCellCycleModel.
Definition at line 58 of file AbstractCellCycleModel.cpp.
References mBirthTime.
Referenced by TissueCell::SetBirthTime().
| void AbstractCellCycleModel::SetCell | ( | TissueCell * | pCell | ) |
Gives the cell cycle model a pointer to its host cell.
Some cell cycle models pass this pointer to other classes (e.g. WntConcentration), which use this information to determine other information based upon the location of the cell (e.g. the Wnt concentration at this location).
- Parameters:
-
pCell pointer to the cell
Definition at line 47 of file AbstractCellCycleModel.cpp.
References mpCell.
Referenced by TissueCell::CommonCopy(), TissueCell::SetCellCycleModel(), and TissueCell::TissueCell().
| void AbstractCellCycleModel::SetDimension | ( | unsigned | dimension | ) |
Set the spatial dimension.
- Parameters:
-
dimension
Definition at line 121 of file AbstractCellCycleModel.cpp.
References mDimension.
Referenced by SingleOdeWntCellCycleModel::SingleOdeWntCellCycleModel().
| virtual void AbstractCellCycleModel::UpdateCellCyclePhase | ( | ) | [pure virtual] |
This method must be implemented by subclasses in order to set the phase the cell cycle model is currently in. It is called from ReadyToDivide() just prior to deciding whether to divide the cell based on how far through the cell cycle it is, i.e. whether it has completed M, G1, S and G2 phases.
Implemented in SimpleWntCellCycleModel, SingleOdeWntCellCycleModel, AbstractOdeBasedCellCycleModel, and AbstractSimpleCellCycleModel.
Referenced by ReadyToDivide().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented in AbstractVanLeeuwen2009WntSwatCellCycleModel, AbstractWntOdeBasedCellCycleModel, SimpleWntCellCycleModel, SingleOdeWntCellCycleModel, StochasticWntCellCycleModel, VanLeeuwen2009WntSwatCellCycleModelHypothesisOne, VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo, WntCellCycleModel, AbstractOdeBasedCellCycleModel, AbstractOdeBasedCellCycleModelWithStoppingEvent, AbstractSimpleCellCycleModel, AbstractSimpleGenerationBasedCellCycleModel, FixedDurationGenerationBasedCellCycleModel, StochasticDurationGenerationBasedCellCycleModel, and TysonNovakCellCycleModel.
Definition at line 56 of file AbstractCellCycleModel.hpp.
Member Data Documentation
double AbstractCellCycleModel::mBirthTime [protected] |
The time that the cell began to split from its parent (i.e. beginning of M phase NOT the end)
Definition at line 105 of file AbstractCellCycleModel.hpp.
Referenced by GetAge(), GetBirthTime(), SingleOdeWntCellCycleModel::Initialise(), AbstractSimpleCellCycleModel::ResetForDivision(), AbstractOdeBasedCellCycleModel::ResetForDivision(), serialize(), SetBirthTime(), and AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase().
CellCyclePhase AbstractCellCycleModel::mCurrentCellCyclePhase [protected] |
The phase of the cell cycle that this model is in (specified in CellCyclePhases.hpp)
Definition at line 108 of file AbstractCellCycleModel.hpp.
Referenced by GetCurrentCellCyclePhase(), ReadyToDivide(), ResetForDivision(), serialize(), AbstractSimpleCellCycleModel::UpdateCellCyclePhase(), and AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase().
unsigned AbstractCellCycleModel::mDimension [protected] |
Spatial dimension being used in simulation (defaults to 0, set with SetDimension)
Definition at line 124 of file AbstractCellCycleModel.hpp.
Referenced by AbstractWntOdeBasedCellCycleModel::AbstractWntOdeBasedCellCycleModel(), GetDimension(), SimpleWntCellCycleModel::GetWntLevel(), AbstractWntOdeBasedCellCycleModel::GetWntLevel(), SimpleWntCellCycleModel::GetWntType(), serialize(), and SetDimension().
double AbstractCellCycleModel::mG1Duration [protected] |
How long the G1 phase lasts for. Not necessarily a fixed value.
Definition at line 114 of file AbstractCellCycleModel.hpp.
Referenced by GetG1Duration(), AbstractOdeBasedCellCycleModel::ResetForDivision(), serialize(), StochasticDurationGenerationBasedCellCycleModel::SetG1Duration(), AbstractSimpleCellCycleModel::SetG1Duration(), SimpleWntCellCycleModel::SetG1Duration(), AbstractSimpleCellCycleModel::UpdateCellCyclePhase(), and AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase().
TissueCell* AbstractCellCycleModel::mpCell [protected] |
The cell that this model is associated with.
Definition at line 99 of file AbstractCellCycleModel.hpp.
Referenced by WntCellCycleModel::ChangeCellProliferativeTypeDueToCurrentBetaCateninLevel(), SingleOdeWntCellCycleModel::ChangeCellProliferativeTypeDueToCurrentBetaCateninLevel(), AbstractVanLeeuwen2009WntSwatCellCycleModel::ChangeCellProliferativeTypeDueToCurrentBetaCateninLevel(), GetCell(), SimpleWntCellCycleModel::GetWntLevel(), AbstractWntOdeBasedCellCycleModel::GetWntLevel(), WntCellCycleModel::Initialise(), SingleOdeWntCellCycleModel::Initialise(), AbstractVanLeeuwen2009WntSwatCellCycleModel::Initialise(), TysonNovakCellCycleModel::InitialiseDaughterCell(), AbstractSimpleGenerationBasedCellCycleModel::InitialiseDaughterCell(), SimpleWntCellCycleModel::InitialiseDaughterCell(), ReadyToDivide(), AbstractSimpleGenerationBasedCellCycleModel::ResetForDivision(), SetCell(), StochasticDurationGenerationBasedCellCycleModel::SetG1Duration(), AbstractSimpleCellCycleModel::SetG1Duration(), SimpleWntCellCycleModel::SetG1Duration(), WntCellCycleModel::SolveOdeToTime(), AbstractVanLeeuwen2009WntSwatCellCycleModel::SolveOdeToTime(), SingleOdeWntCellCycleModel::UpdateBetaCateninLevel(), AbstractSimpleCellCycleModel::UpdateCellCyclePhase(), SimpleWntCellCycleModel::UpdateCellCyclePhase(), and AbstractWntOdeBasedCellCycleModel::UpdateCellProliferativeType().
bool AbstractCellCycleModel::mReadyToDivide [protected] |
Whether the cell is currently ready to undergo division.
Definition at line 119 of file AbstractCellCycleModel.hpp.
Referenced by ReadyToDivide(), ResetForDivision(), and serialize().
The documentation for this class was generated from the following files:
- /tmp/release_2.0/cell_based/src/tissue/cell/cycle/AbstractCellCycleModel.hpp
- /tmp/release_2.0/cell_based/src/tissue/cell/cycle/AbstractCellCycleModel.cpp
 1.6.2
1.6.2