StochasticOxygenBasedCellCycleModel Class Reference
#include <StochasticOxygenBasedCellCycleModel.hpp>
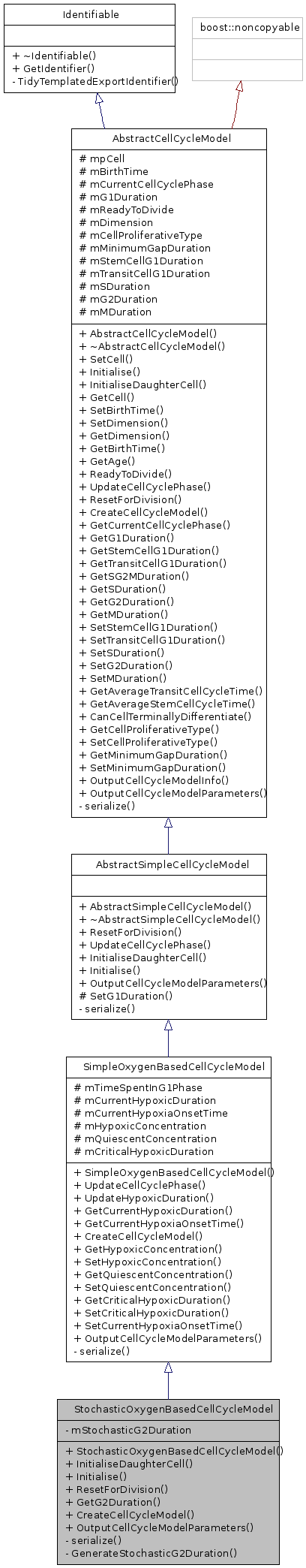
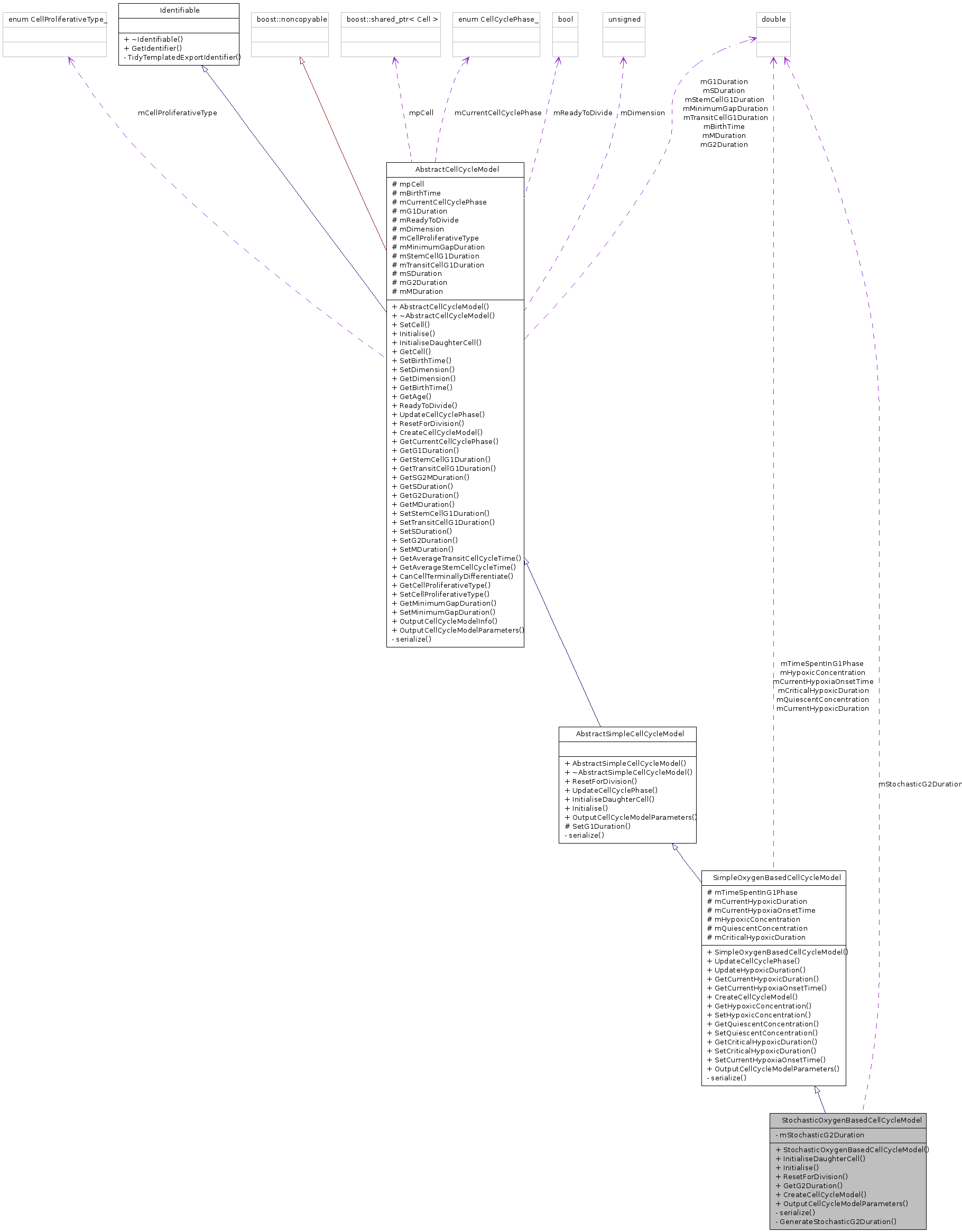
Public Member Functions | |
| StochasticOxygenBasedCellCycleModel () | |
| void | InitialiseDaughterCell () |
| void | Initialise () |
| void | ResetForDivision () |
| double | GetG2Duration () |
| AbstractCellCycleModel * | CreateCellCycleModel () |
| virtual void | OutputCellCycleModelParameters (out_stream &rParamsFile) |
Private Member Functions | |
| template<class Archive> | |
| void | serialize (Archive &archive, const unsigned int version) |
| void | GenerateStochasticG2Duration () |
Private Attributes | |
| double | mStochasticG2Duration |
Friends | |
| class | boost::serialization::access |
Detailed Description
Stochastic oxygen-based cell-cycle model.A simple oxygen-dependent cell-cycle model that inherits from SimpleOxygenBasedCellCycleModel and in addition spends a random duration in G2 phase.
Definition at line 41 of file StochasticOxygenBasedCellCycleModel.hpp.
Constructor & Destructor Documentation
| StochasticOxygenBasedCellCycleModel::StochasticOxygenBasedCellCycleModel | ( | ) |
Constructor.
Definition at line 32 of file StochasticOxygenBasedCellCycleModel.cpp.
Referenced by CreateCellCycleModel().
Member Function Documentation
| void StochasticOxygenBasedCellCycleModel::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Archive the cell-cycle model.
- Parameters:
-
archive the archive version the current version of this class
Reimplemented from SimpleOxygenBasedCellCycleModel.
Definition at line 47 of file StochasticOxygenBasedCellCycleModel.hpp.
References RandomNumberGenerator::Instance(), and mStochasticG2Duration.
| void StochasticOxygenBasedCellCycleModel::GenerateStochasticG2Duration | ( | ) | [private] |
Stochastically set the G2 duration. Called on cell creation at the start of a simulation, and for both parent and daughter cells at cell division.
Definition at line 37 of file StochasticOxygenBasedCellCycleModel.cpp.
References AbstractCellCycleModel::GetG2Duration(), RandomNumberGenerator::Instance(), AbstractCellCycleModel::mMinimumGapDuration, mStochasticG2Duration, and RandomNumberGenerator::NormalRandomDeviate().
Referenced by Initialise(), InitialiseDaughterCell(), and ResetForDivision().
| void StochasticOxygenBasedCellCycleModel::InitialiseDaughterCell | ( | ) | [virtual] |
Overridden InitialiseDaughterCell() method.
Reimplemented from AbstractSimpleCellCycleModel.
Definition at line 53 of file StochasticOxygenBasedCellCycleModel.cpp.
References GenerateStochasticG2Duration(), and AbstractSimpleCellCycleModel::InitialiseDaughterCell().
| void StochasticOxygenBasedCellCycleModel::Initialise | ( | void | ) | [virtual] |
Initialise the cell-cycle model at the start of a simulation.
Reimplemented from AbstractSimpleCellCycleModel.
Definition at line 59 of file StochasticOxygenBasedCellCycleModel.cpp.
References GenerateStochasticG2Duration(), and AbstractSimpleCellCycleModel::Initialise().
| void StochasticOxygenBasedCellCycleModel::ResetForDivision | ( | ) | [virtual] |
Overridden ResetForDivision() method.
Reimplemented from AbstractSimpleCellCycleModel.
Definition at line 65 of file StochasticOxygenBasedCellCycleModel.cpp.
References GenerateStochasticG2Duration(), and AbstractSimpleCellCycleModel::ResetForDivision().
| double StochasticOxygenBasedCellCycleModel::GetG2Duration | ( | ) | [virtual] |
- Returns:
- mStochasticG2Duration.
Reimplemented from AbstractCellCycleModel.
Definition at line 71 of file StochasticOxygenBasedCellCycleModel.cpp.
References mStochasticG2Duration.
| AbstractCellCycleModel * StochasticOxygenBasedCellCycleModel::CreateCellCycleModel | ( | ) | [virtual] |
Overridden builder method to create new copies of this cell-cycle model.
Reimplemented from SimpleOxygenBasedCellCycleModel.
Definition at line 76 of file StochasticOxygenBasedCellCycleModel.cpp.
References AbstractCellCycleModel::mBirthTime, AbstractCellCycleModel::mCellProliferativeType, SimpleOxygenBasedCellCycleModel::mCriticalHypoxicDuration, SimpleOxygenBasedCellCycleModel::mCurrentHypoxiaOnsetTime, AbstractCellCycleModel::mDimension, AbstractCellCycleModel::mG2Duration, SimpleOxygenBasedCellCycleModel::mHypoxicConcentration, AbstractCellCycleModel::mMDuration, AbstractCellCycleModel::mMinimumGapDuration, SimpleOxygenBasedCellCycleModel::mQuiescentConcentration, AbstractCellCycleModel::mSDuration, AbstractCellCycleModel::mStemCellG1Duration, AbstractCellCycleModel::mTransitCellG1Duration, AbstractCellCycleModel::SetBirthTime(), AbstractCellCycleModel::SetCellProliferativeType(), SimpleOxygenBasedCellCycleModel::SetCriticalHypoxicDuration(), SimpleOxygenBasedCellCycleModel::SetCurrentHypoxiaOnsetTime(), AbstractCellCycleModel::SetDimension(), AbstractCellCycleModel::SetG2Duration(), SimpleOxygenBasedCellCycleModel::SetHypoxicConcentration(), AbstractCellCycleModel::SetMDuration(), AbstractCellCycleModel::SetMinimumGapDuration(), SimpleOxygenBasedCellCycleModel::SetQuiescentConcentration(), AbstractCellCycleModel::SetSDuration(), AbstractCellCycleModel::SetStemCellG1Duration(), AbstractCellCycleModel::SetTransitCellG1Duration(), and StochasticOxygenBasedCellCycleModel().
| void StochasticOxygenBasedCellCycleModel::OutputCellCycleModelParameters | ( | out_stream & | rParamsFile | ) | [virtual] |
Outputs cell cycle model parameters to file.
- Parameters:
-
rParamsFile the file stream to which the parameters are output
Reimplemented from SimpleOxygenBasedCellCycleModel.
Definition at line 111 of file StochasticOxygenBasedCellCycleModel.cpp.
References SimpleOxygenBasedCellCycleModel::OutputCellCycleModelParameters().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented from SimpleOxygenBasedCellCycleModel.
Definition at line 45 of file StochasticOxygenBasedCellCycleModel.hpp.
Member Data Documentation
double StochasticOxygenBasedCellCycleModel::mStochasticG2Duration [private] |
The duration of the G2 phase, set stochastically.
Definition at line 61 of file StochasticOxygenBasedCellCycleModel.hpp.
Referenced by GenerateStochasticG2Duration(), GetG2Duration(), and serialize().
The documentation for this class was generated from the following files:
- cell_based/src/population/cell/cycle/StochasticOxygenBasedCellCycleModel.hpp
- cell_based/src/population/cell/cycle/StochasticOxygenBasedCellCycleModel.cpp
 1.5.5
1.5.5