AbstractCellPopulation< DIM > Class Template Reference
#include <AbstractCellPopulation.hpp>
Inherits Identifiable.
Inherited by AbstractOffLatticeCellPopulation< DIM >, and AbstractOnLatticeCellPopulation< DIM >.
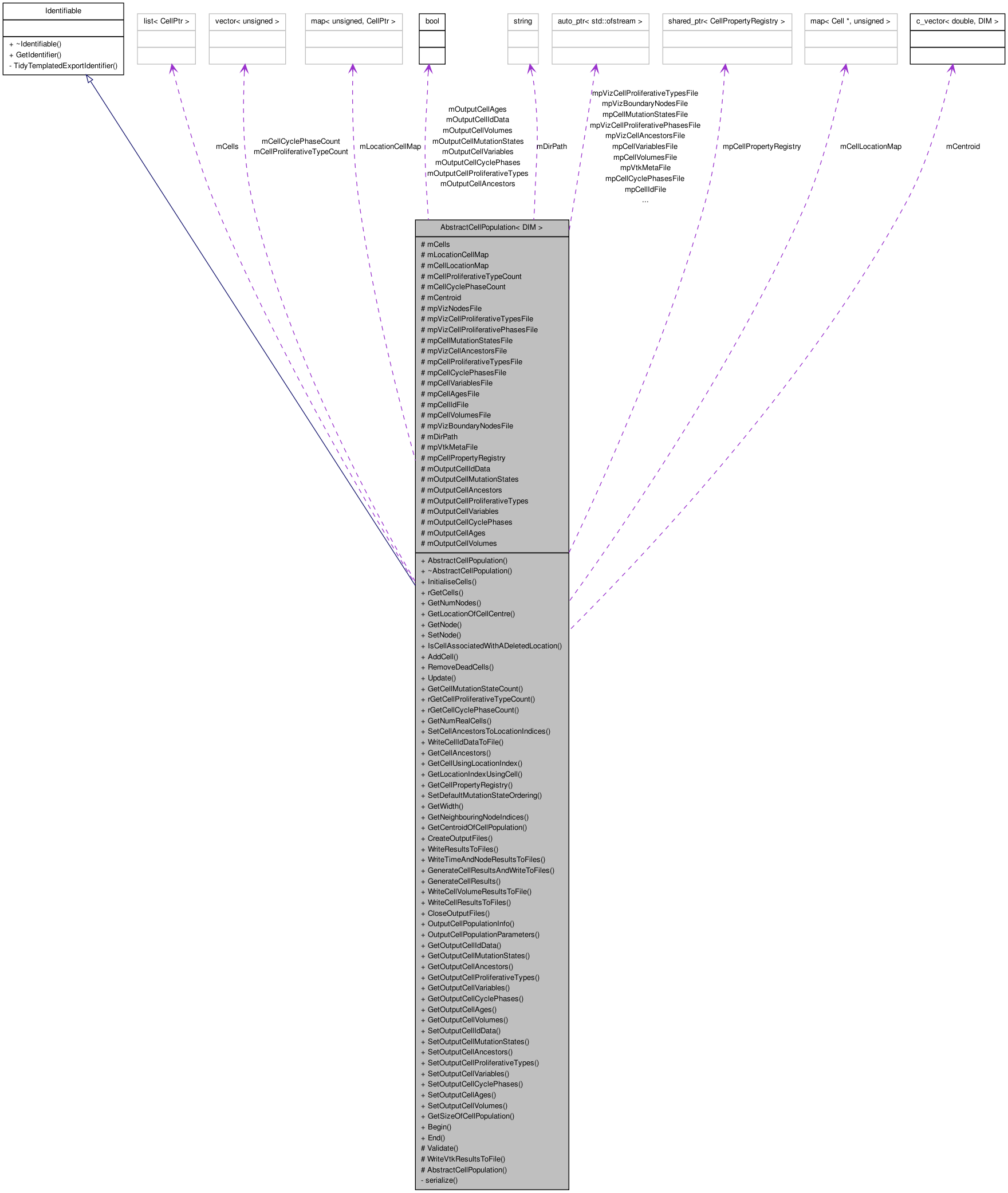
Detailed Description
template<unsigned DIM>
class AbstractCellPopulation< DIM >
An abstract facade class encapsulating a cell population.
Contains a group of cells and associated methods.
Definition at line 66 of file AbstractCellPopulation.hpp.
Constructor & Destructor Documentation
| AbstractCellPopulation< DIM >::AbstractCellPopulation | ( | ) | [inline, protected] |
Constructor for use by archiving only. Please use the other constructor.
Doesn't take in cells, since these are dealt with by the serialize method.
Definition at line 206 of file AbstractCellPopulation.hpp.
| AbstractCellPopulation< DIM >::AbstractCellPopulation | ( | std::vector< CellPtr > & | rCells, | |
| const std::vector< unsigned > | locationIndices = std::vector<unsigned>() | |||
| ) | [inline] |
AbstractCellPopulation Constructor.
- Note:
- Warning: the passed-in vector of cells will be emptied, even if the constructor throws an exception!
- Parameters:
-
rCells a vector of cells. Copies of the cells will be stored in the cell population, and the passed-in vector cleared. locationIndices an optional vector of location indices that correspond to real cells
Definition at line 34 of file AbstractCellPopulation.cpp.
References EXCEPTION, AbstractCellPopulation< DIM >::mCellCyclePhaseCount, AbstractCellPopulation< DIM >::mCellLocationMap, AbstractCellPopulation< DIM >::mCellProliferativeTypeCount, AbstractCellPopulation< DIM >::mCells, AbstractCellPopulation< DIM >::mLocationCellMap, and AbstractCellPopulation< DIM >::mpCellPropertyRegistry.
| AbstractCellPopulation< DIM >::~AbstractCellPopulation | ( | ) | [inline, virtual] |
Base class with virtual methods needs a virtual destructor.
Definition at line 99 of file AbstractCellPopulation.cpp.
Member Function Documentation
| virtual CellPtr AbstractCellPopulation< DIM >::AddCell | ( | CellPtr | pNewCell, | |
| const c_vector< double, DIM > & | rCellDivisionVector, | |||
| CellPtr | pParentCell = CellPtr() | |||
| ) | [pure virtual] |
Add a new cell to the cell population.
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
pNewCell the cell to add rCellDivisionVector a vector providing information regarding how the cell division should occur (for cell-centre cell populations, this vector is the position of the daughter cell; for vertex cell populations it can be used by any subclass of CellBasedSimulation to as a means of dictating the axis along which the parent cell divides) pParentCell pointer to a parent cell (if required)
- Returns:
- address of cell as it appears in the cell list (internal of this method uses a copy constructor along the way).
Implemented in AbstractCentreBasedCellPopulation< DIM >, CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, MeshBasedCellPopulationWithGhostNodes< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
| AbstractCellPopulation< DIM >::Iterator AbstractCellPopulation< DIM >::Begin | ( | ) | [inline] |
- Returns:
- iterator pointing to the first cell in the cell population
Definition at line 788 of file AbstractCellPopulation.hpp.
References AbstractCellPopulation< DIM >::mCells.
Referenced by CryptProjectionForce::AddForceContribution(), ChemotacticForce< DIM >::AddForceContribution(), BuskeCompressionForce< DIM >::AddForceContribution(), VertexBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), PottsBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCellPopulation< DIM >::GetCellAncestors(), AbstractCellPopulation< DIM >::GetCentroidOfCellPopulation(), CryptStatistics::GetCryptSection(), CryptProjectionStatistics::GetCryptSection(), NodeBasedCellPopulation< DIM >::GetNeighbouringNodeIndices(), AbstractCellPopulation< DIM >::GetNumRealCells(), AbstractCellPopulation< DIM >::GetSizeOfCellPopulation(), AbstractCellPopulation< DIM >::InitialiseCells(), AbstractCryptStatistics::LabelAllCellsAsHealthy(), AbstractCryptStatistics::LabelSPhaseCells(), AbstractCellPopulation< DIM >::OutputCellPopulationInfo(), AbstractCellPopulation< DIM >::SetCellAncestorsToLocationIndices(), CellwiseDataGradient< DIM >::SetupGradients(), SloughingCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), RadialSloughingCellKiller::TestAndLabelCellsForApoptosisOrDeath(), RandomCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), PlaneBasedCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), OxygenBasedCellKiller< SPACE_DIM >::TestAndLabelCellsForApoptosisOrDeath(), CaBasedCellPopulation< DIM >::UpdateCellLocations(), CryptProjectionForce::UpdateNode3dLocationMap(), NodeBasedCellPopulationWithBuskeUpdate< DIM >::UpdateNodeLocations(), AbstractCentreBasedCellPopulation< DIM >::UpdateNodeLocations(), VertexBasedCellPopulation< DIM >::Validate(), PottsBasedCellPopulation< DIM >::Validate(), NodeBasedCellPopulation< DIM >::Validate(), MeshBasedCellPopulationWithGhostNodes< DIM >::Validate(), MeshBasedCellPopulation< DIM >::Validate(), CaBasedCellPopulation< DIM >::Validate(), AbstractCellPopulation< DIM >::WriteCellIdDataToFile(), VertexBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), PottsBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), NodeBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), CaBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
| void AbstractCellPopulation< DIM >::CloseOutputFiles | ( | ) | [inline, virtual] |
Close any output files.
Reimplemented in MeshBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Definition at line 329 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellAges, AbstractCellPopulation< DIM >::mOutputCellAncestors, AbstractCellPopulation< DIM >::mOutputCellCyclePhases, AbstractCellPopulation< DIM >::mOutputCellIdData, AbstractCellPopulation< DIM >::mOutputCellMutationStates, AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes, AbstractCellPopulation< DIM >::mOutputCellVariables, AbstractCellPopulation< DIM >::mOutputCellVolumes, AbstractCellPopulation< DIM >::mpCellAgesFile, AbstractCellPopulation< DIM >::mpCellCyclePhasesFile, AbstractCellPopulation< DIM >::mpCellIdFile, AbstractCellPopulation< DIM >::mpCellMutationStatesFile, AbstractCellPopulation< DIM >::mpCellProliferativeTypesFile, AbstractCellPopulation< DIM >::mpCellVariablesFile, AbstractCellPopulation< DIM >::mpCellVolumesFile, AbstractCellPopulation< DIM >::mpVizBoundaryNodesFile, AbstractCellPopulation< DIM >::mpVizCellAncestorsFile, AbstractCellPopulation< DIM >::mpVizCellProliferativePhasesFile, AbstractCellPopulation< DIM >::mpVizCellProliferativeTypesFile, AbstractCellPopulation< DIM >::mpVizNodesFile, and AbstractCellPopulation< DIM >::mpVtkMetaFile.
| void AbstractCellPopulation< DIM >::CreateOutputFiles | ( | const std::string & | rDirectory, | |
| bool | cleanOutputDirectory | |||
| ) | [inline, virtual] |
Use an output file handler to create output files for visualizer and post-processing.
- Parameters:
-
rDirectory pathname of the output directory, relative to where Chaste output is stored cleanOutputDirectory whether to delete the contents of the output directory prior to output file creation
Reimplemented in MeshBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Definition at line 260 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mDirPath, AbstractCellPopulation< DIM >::mOutputCellAges, AbstractCellPopulation< DIM >::mOutputCellAncestors, AbstractCellPopulation< DIM >::mOutputCellCyclePhases, AbstractCellPopulation< DIM >::mOutputCellIdData, AbstractCellPopulation< DIM >::mOutputCellMutationStates, AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes, AbstractCellPopulation< DIM >::mOutputCellVariables, AbstractCellPopulation< DIM >::mOutputCellVolumes, AbstractCellPopulation< DIM >::mpCellAgesFile, AbstractCellPopulation< DIM >::mpCellCyclePhasesFile, AbstractCellPopulation< DIM >::mpCellIdFile, AbstractCellPopulation< DIM >::mpCellMutationStatesFile, AbstractCellPopulation< DIM >::mpCellProliferativeTypesFile, AbstractCellPopulation< DIM >::mpCellPropertyRegistry, AbstractCellPopulation< DIM >::mpCellVariablesFile, AbstractCellPopulation< DIM >::mpCellVolumesFile, AbstractCellPopulation< DIM >::mpVizBoundaryNodesFile, AbstractCellPopulation< DIM >::mpVizCellAncestorsFile, AbstractCellPopulation< DIM >::mpVizCellProliferativePhasesFile, AbstractCellPopulation< DIM >::mpVizCellProliferativeTypesFile, AbstractCellPopulation< DIM >::mpVizNodesFile, AbstractCellPopulation< DIM >::mpVtkMetaFile, OutputFileHandler::OpenOutputFile(), and AbstractCellPopulation< DIM >::SetDefaultMutationStateOrdering().
| AbstractCellPopulation< DIM >::Iterator AbstractCellPopulation< DIM >::End | ( | ) | [inline] |
- Returns:
- iterator pointing to one past the last cell in the cell population
Definition at line 794 of file AbstractCellPopulation.hpp.
References AbstractCellPopulation< DIM >::mCells.
Referenced by CryptProjectionForce::AddForceContribution(), ChemotacticForce< DIM >::AddForceContribution(), BuskeCompressionForce< DIM >::AddForceContribution(), VertexBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), PottsBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCellPopulation< DIM >::GetCellAncestors(), AbstractCellPopulation< DIM >::GetCentroidOfCellPopulation(), CryptStatistics::GetCryptSection(), CryptProjectionStatistics::GetCryptSection(), NodeBasedCellPopulation< DIM >::GetNeighbouringNodeIndices(), AbstractCellPopulation< DIM >::GetNumRealCells(), AbstractCellPopulation< DIM >::GetSizeOfCellPopulation(), AbstractCellPopulation< DIM >::InitialiseCells(), AbstractCryptStatistics::LabelAllCellsAsHealthy(), AbstractCryptStatistics::LabelSPhaseCells(), AbstractCellPopulation< DIM >::OutputCellPopulationInfo(), AbstractCellPopulation< DIM >::SetCellAncestorsToLocationIndices(), CellwiseDataGradient< DIM >::SetupGradients(), SloughingCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), RadialSloughingCellKiller::TestAndLabelCellsForApoptosisOrDeath(), RandomCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), PlaneBasedCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), OxygenBasedCellKiller< SPACE_DIM >::TestAndLabelCellsForApoptosisOrDeath(), CaBasedCellPopulation< DIM >::UpdateCellLocations(), CryptProjectionForce::UpdateNode3dLocationMap(), NodeBasedCellPopulationWithBuskeUpdate< DIM >::UpdateNodeLocations(), AbstractCentreBasedCellPopulation< DIM >::UpdateNodeLocations(), VertexBasedCellPopulation< DIM >::Validate(), PottsBasedCellPopulation< DIM >::Validate(), NodeBasedCellPopulation< DIM >::Validate(), MeshBasedCellPopulationWithGhostNodes< DIM >::Validate(), MeshBasedCellPopulation< DIM >::Validate(), CaBasedCellPopulation< DIM >::Validate(), AbstractCellPopulation< DIM >::WriteCellIdDataToFile(), VertexBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), PottsBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), NodeBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), CaBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
| void AbstractCellPopulation< DIM >::GenerateCellResults | ( | unsigned | locationIndex, | |
| std::vector< unsigned > & | rCellProliferativeTypeCounter, | |||
| std::vector< unsigned > & | rCellCyclePhaseCounter | |||
| ) | [inline, virtual] |
Generate results for a given cell in the current cell population state to output files.
- Parameters:
-
locationIndex location index of the cell rCellProliferativeTypeCounter cell type counter rCellCyclePhaseCounter cell cycle phase counter
Reimplemented in AbstractCentreBasedCellPopulation< DIM >, and CaBasedCellPopulation< DIM >.
Definition at line 377 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::GetLocationOfCellCentre(), CellPropertyCollection::GetProperty(), AbstractCellPopulation< DIM >::mLocationCellMap, AbstractCellPopulation< DIM >::mOutputCellAges, AbstractCellPopulation< DIM >::mOutputCellAncestors, AbstractCellPopulation< DIM >::mOutputCellCyclePhases, AbstractCellPopulation< DIM >::mOutputCellMutationStates, AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes, AbstractCellPopulation< DIM >::mOutputCellVariables, AbstractCellPopulation< DIM >::mpCellAgesFile, AbstractCellPopulation< DIM >::mpCellVariablesFile, AbstractCellPopulation< DIM >::mpVizCellAncestorsFile, AbstractCellPopulation< DIM >::mpVizCellProliferativePhasesFile, AbstractCellPopulation< DIM >::mpVizCellProliferativeTypesFile, NEVER_REACHED, and UNSIGNED_UNSET.
Referenced by VertexBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), and PottsBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles().
| virtual void AbstractCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles | ( | ) | [pure virtual] |
Calls GenerateCellResults() on each cell then calls WriteCellResultsToFiles().
Implemented in AbstractCentreBasedCellPopulation< DIM >, CaBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, and VertexBasedCellPopulation< DIM >.
Referenced by AbstractCellPopulation< DIM >::WriteResultsToFiles().
| std::set< unsigned > AbstractCellPopulation< DIM >::GetCellAncestors | ( | ) | [inline] |
Loops over cells and makes a list of the ancestors that are part of the cell population.
- Returns:
- remaining_ancestors The size of this set tells you how many clonal populations remain.
Definition at line 139 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), and AbstractCellPopulation< DIM >::End().
| std::vector< unsigned > AbstractCellPopulation< DIM >::GetCellMutationStateCount | ( | ) | [inline] |
Find out how many cells of each mutation state there are
- Returns:
- The number of cells of each mutation state (evaluated at each visualizer output) [0] = healthy count [1] = APC one hit [2] = APC two hit [3] = beta catenin one hit
Definition at line 150 of file AbstractCellPopulation.cpp.
References EXCEPTION, AbstractCellPopulation< DIM >::mOutputCellMutationStates, AbstractCellPopulation< DIM >::mpCellPropertyRegistry, and AbstractCellPopulation< DIM >::SetDefaultMutationStateOrdering().
Referenced by AbstractCellPopulation< DIM >::WriteCellResultsToFiles().
| boost::shared_ptr< CellPropertyRegistry > AbstractCellPopulation< DIM >::GetCellPropertyRegistry | ( | ) | [inline] |
- Returns:
- registry of cell properties used in this cell population.
Definition at line 219 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mpCellPropertyRegistry.
Referenced by AbstractCellPopulation< DIM >::SetDefaultMutationStateOrdering().
| CellPtr AbstractCellPopulation< DIM >::GetCellUsingLocationIndex | ( | unsigned | index | ) | [inline] |
Get the cell corresponding to a given location index.
Currently assumes there is one cell for each location index, and they are ordered identically in their vectors. An assertion fails if not.
- Parameters:
-
index the location index
- Returns:
- the cell.
Definition at line 196 of file AbstractCellPopulation.cpp.
References EXCEPTION, and AbstractCellPopulation< DIM >::mLocationCellMap.
Referenced by NagaiHondaForce< DIM >::AddForceContribution(), CryptProjectionForce::CalculateForceBetweenNodes(), GeneralisedLinearSpringForce< DIM >::CalculateForceBetweenNodes(), MeshBasedCellPopulation< DIM >::CheckCellPointers(), AdhesionPottsUpdateRule< DIM >::EvaluateHamiltonianContribution(), VertexBasedCellPopulation< DIM >::GetDampingConstant(), AbstractCentreBasedCellPopulation< DIM >::GetDampingConstant(), CellwiseDataGradient< DIM >::SetupGradients(), TargetedCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), and LinearSpringWithVariableSpringConstantsForce< DIM >::VariableSpringConstantMultiplicationFactor().
| c_vector< double, DIM > AbstractCellPopulation< DIM >::GetCentroidOfCellPopulation | ( | ) | [inline] |
Returns the centroid of the cell population.
Definition at line 240 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), AbstractCellPopulation< DIM >::End(), AbstractCellPopulation< DIM >::GetLocationOfCellCentre(), AbstractCellPopulation< DIM >::GetNumRealCells(), and AbstractCellPopulation< DIM >::mCentroid.
Referenced by AbstractCellPopulation< DIM >::GetSizeOfCellPopulation().
| unsigned AbstractCellPopulation< DIM >::GetLocationIndexUsingCell | ( | CellPtr | pCell | ) | [inline] |
Get the location index corresponding to a given cell.
Currently assumes there is one cell for each location index, and they are ordered identically in their vectors. An assertion fails if not.
- Parameters:
-
pCell the cell
- Returns:
- the location index.
Definition at line 213 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mCellLocationMap.
Referenced by NodeBasedCellPopulation< DIM >::AddCell(), CryptProjectionForce::AddForceContribution(), ChemotacticForce< DIM >::AddForceContribution(), BuskeCompressionForce< DIM >::AddForceContribution(), CryptSimulation1d::CalculateCellDivisionVector(), VertexBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), PottsBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), NodeBasedCellPopulation< DIM >::GetNeighbouringNodeIndices(), CaBasedCellPopulation< DIM >::RemoveDeadCells(), CellwiseDataGradient< DIM >::SetupGradients(), CaBasedCellPopulation< DIM >::UpdateCellLocations(), CryptProjectionForce::UpdateNode3dLocationMap(), VertexBasedCellPopulation< DIM >::Validate(), PottsBasedCellPopulation< DIM >::Validate(), MeshBasedCellPopulation< DIM >::Validate(), VertexBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), PottsBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), NodeBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), CaBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), VertexBasedCellPopulation< DIM >::WriteResultsToFiles(), PottsBasedCellPopulation< DIM >::WriteResultsToFiles(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
| virtual c_vector<double, DIM> AbstractCellPopulation< DIM >::GetLocationOfCellCentre | ( | CellPtr | pCell | ) | [pure virtual] |
Find where a given cell is in space.
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
pCell the cell
- Returns:
- the location of the cell
Implemented in AbstractCentreBasedCellPopulation< DIM >, CaBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, and VertexBasedCellPopulation< DIM >.
Referenced by AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GetCentroidOfCellPopulation(), AbstractCellPopulation< DIM >::GetSizeOfCellPopulation(), SloughingCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), RadialSloughingCellKiller::TestAndLabelCellsForApoptosisOrDeath(), PlaneBasedCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath(), CryptProjectionForce::UpdateNode3dLocationMap(), and AbstractCellPopulation< DIM >::WriteCellIdDataToFile().
| virtual std::set<unsigned> AbstractCellPopulation< DIM >::GetNeighbouringNodeIndices | ( | unsigned | index | ) | [pure virtual] |
Given a node index, returns the set of neighbouring node indices.
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
index the node index
- Returns:
- the set of neighbouring node indices.
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
| virtual Node<DIM>* AbstractCellPopulation< DIM >::GetNode | ( | unsigned | index | ) | [pure virtual] |
Get a pointer to the node with a given index.
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
index global index of the specified node
- Returns:
- a pointer to the node with a given index.
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Referenced by RepulsionForce< DIM >::AddForceContribution(), BuskeCompressionForce< DIM >::AddForceContribution(), CryptProjectionForce::CalculateForceBetweenNodes(), GeneralisedLinearSpringForce< DIM >::CalculateForceBetweenNodes(), BuskeElasticForce< DIM >::CalculateForceBetweenNodes(), BuskeAdhesiveForce< DIM >::CalculateForceBetweenNodes(), AbstractCentreBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCentreBasedCellPopulation< DIM >::GetNodeCorrespondingToCell(), AbstractCentreBasedCellPopulation< DIM >::UpdateNodeLocations(), AbstractCentreBasedCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles(), and AbstractCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles().
| virtual unsigned AbstractCellPopulation< DIM >::GetNumNodes | ( | ) | [pure virtual] |
As this method is pure virtual, it must be overridden in subclasses.
- Returns:
- the number of nodes in the cell population.
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Referenced by AbstractCentreBasedCellPopulation< DIM >::AddCell(), AbstractCentreBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCentreBasedCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles(), and AbstractCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles().
| unsigned AbstractCellPopulation< DIM >::GetNumRealCells | ( | ) | [inline] |
Get the number of real cells.
Definition at line 119 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), and AbstractCellPopulation< DIM >::End().
Referenced by AbstractCellPopulation< DIM >::GetCentroidOfCellPopulation(), and TargetedCellKiller< DIM >::TestAndLabelCellsForApoptosisOrDeath().
| bool AbstractCellPopulation< DIM >::GetOutputCellAges | ( | ) | [inline] |
- Returns:
- mOutputCellAges
Definition at line 767 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellAges.
| bool AbstractCellPopulation< DIM >::GetOutputCellAncestors | ( | ) | [inline] |
- Returns:
- mOutputCellAncestors
Definition at line 743 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellAncestors.
| bool AbstractCellPopulation< DIM >::GetOutputCellCyclePhases | ( | ) | [inline] |
- Returns:
- mOutputCellCyclePhases
Definition at line 761 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellCyclePhases.
| bool AbstractCellPopulation< DIM >::GetOutputCellIdData | ( | ) | [inline] |
- Returns:
- mOutputCellIdData
Definition at line 731 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellIdData.
| bool AbstractCellPopulation< DIM >::GetOutputCellMutationStates | ( | ) | [inline] |
- Returns:
- mOutputCellMutationStates
Definition at line 737 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellMutationStates.
| bool AbstractCellPopulation< DIM >::GetOutputCellProliferativeTypes | ( | ) | [inline] |
- Returns:
- mOutputCellProliferativeTypes
Definition at line 749 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes.
| bool AbstractCellPopulation< DIM >::GetOutputCellVariables | ( | ) | [inline] |
- Returns:
- mOutputCellVariables
Definition at line 755 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellVariables.
| bool AbstractCellPopulation< DIM >::GetOutputCellVolumes | ( | ) | [inline] |
- Returns:
- mOutputCellVolumes
Definition at line 773 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellVolumes.
| c_vector< double, DIM > AbstractCellPopulation< DIM >::GetSizeOfCellPopulation | ( | ) | [inline] |
- Returns:
- The width (maximum distance to centroid) of the cell population in each dimension
Definition at line 831 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), AbstractCellPopulation< DIM >::End(), AbstractCellPopulation< DIM >::GetCentroidOfCellPopulation(), and AbstractCellPopulation< DIM >::GetLocationOfCellCentre().
| virtual double AbstractCellPopulation< DIM >::GetWidth | ( | const unsigned & | rDimension | ) | [pure virtual] |
Calculate the 'width' of any dimension of the cell population.
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
rDimension a dimension (0,1 or 2)
- Returns:
- The maximum distance between any nodes in this dimension.
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
| void AbstractCellPopulation< DIM >::InitialiseCells | ( | ) | [inline] |
Initialise each cell's cell-cycle model.
Definition at line 104 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), and AbstractCellPopulation< DIM >::End().
| virtual bool AbstractCellPopulation< DIM >::IsCellAssociatedWithADeletedLocation | ( | CellPtr | pCell | ) | [pure virtual] |
Helper method for establishing if a cell is real.
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
pCell the cell
- Returns:
- whether a given cell is associated with a deleted node (cell-centre models) or element (vertex models).
Implemented in AbstractCentreBasedCellPopulation< DIM >, CaBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, and VertexBasedCellPopulation< DIM >.
Referenced by AbstractCellPopulation< DIM >::Iterator::IsRealCell().
| void AbstractCellPopulation< DIM >::OutputCellPopulationInfo | ( | out_stream & | rParamsFile | ) | [inline] |
Outputs CellPopulation used in the simulation to file and then calls OutputCellPopulationParameters to output all relevant parameters.
- Parameters:
-
rParamsFile the file stream to which the parameters are output
Definition at line 673 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), AbstractCellPopulation< DIM >::End(), Identifiable::GetIdentifier(), and AbstractCellPopulation< DIM >::OutputCellPopulationParameters().
| void AbstractCellPopulation< DIM >::OutputCellPopulationParameters | ( | out_stream & | rParamsFile | ) | [inline, pure virtual] |
Outputs CellPopulation parameters to file
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
rParamsFile the file stream to which the parameters are output
Implemented in AbstractCentreBasedCellPopulation< DIM >, AbstractOffLatticeCellPopulation< DIM >, AbstractOnLatticeCellPopulation< DIM >, CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, MeshBasedCellPopulationWithGhostNodes< DIM >, NodeBasedCellPopulation< DIM >, NodeBasedCellPopulationWithBuskeUpdate< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Definition at line 714 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellAges, AbstractCellPopulation< DIM >::mOutputCellAncestors, AbstractCellPopulation< DIM >::mOutputCellCyclePhases, AbstractCellPopulation< DIM >::mOutputCellIdData, AbstractCellPopulation< DIM >::mOutputCellMutationStates, AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes, AbstractCellPopulation< DIM >::mOutputCellVariables, and AbstractCellPopulation< DIM >::mOutputCellVolumes.
Referenced by AbstractCellPopulation< DIM >::OutputCellPopulationInfo().
| virtual unsigned AbstractCellPopulation< DIM >::RemoveDeadCells | ( | ) | [pure virtual] |
Remove all cells labelled as dead.
As this method is pure virtual, it must be overridden in subclasses.
- Returns:
- number of cells removed
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
| const std::vector< unsigned > & AbstractCellPopulation< DIM >::rGetCellCyclePhaseCount | ( | ) | const [inline] |
Find out how many cells in each cell cycle phase there are
- Returns:
- The number of cells of each phase (evaluated at each visualizer output) [0] = G_ZERO_PHASE [1] = G_ONE_PHASE [2] = S_PHASE [3] = G_TWO_PHASE [4] = M_PHASE
Definition at line 186 of file AbstractCellPopulation.cpp.
References EXCEPTION, AbstractCellPopulation< DIM >::mCellCyclePhaseCount, and AbstractCellPopulation< DIM >::mOutputCellCyclePhases.
| const std::vector< unsigned > & AbstractCellPopulation< DIM >::rGetCellProliferativeTypeCount | ( | ) | const [inline] |
Find out how many cells of each type there are
- Returns:
- The number of cells of each type (evaluated at each visualizer output) [0] = STEM [1] = TRANSIT [2] = DIFFERENTIATED
Definition at line 176 of file AbstractCellPopulation.cpp.
References EXCEPTION, AbstractCellPopulation< DIM >::mCellProliferativeTypeCount, and AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes.
| std::list< CellPtr > & AbstractCellPopulation< DIM >::rGetCells | ( | ) | [inline] |
- Returns:
- reference to mCells.
Definition at line 113 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mCells.
Referenced by AbstractCellPopulation< DIM >::Iterator::IsAtEnd(), and AbstractCellPopulation< DIM >::Iterator::Iterator().
| void AbstractCellPopulation< DIM >::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Serialize the object and its member variables.
- Parameters:
-
archive the archive version the current version of this class
Reimplemented in AbstractCentreBasedCellPopulation< DIM >, AbstractOffLatticeCellPopulation< DIM >, AbstractOnLatticeCellPopulation< DIM >, CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, MeshBasedCellPopulationWithGhostNodes< DIM >, NodeBasedCellPopulation< DIM >, NodeBasedCellPopulationWithBuskeUpdate< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Definition at line 78 of file AbstractCellPopulation.hpp.
| void AbstractCellPopulation< DIM >::SetCellAncestorsToLocationIndices | ( | ) | [inline] |
Sets the Ancestor index of all the cells at this time to be the same as their location index, can be used to trace clonal populations.
Definition at line 130 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), AbstractCellPopulation< DIM >::End(), and AbstractCellPopulation< DIM >::mCellLocationMap.
| void AbstractCellPopulation< DIM >::SetDefaultMutationStateOrdering | ( | ) | [inline] |
Set a default ordering on mutation states, so that existing tests don't need to specify the old ordering explicitly.
Definition at line 225 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::GetCellPropertyRegistry().
Referenced by AbstractCellPopulation< DIM >::CreateOutputFiles(), and AbstractCellPopulation< DIM >::GetCellMutationStateCount().
| virtual void AbstractCellPopulation< DIM >::SetNode | ( | unsigned | nodeIndex, | |
| ChastePoint< DIM > & | rNewLocation | |||
| ) | [pure virtual] |
Move the node with a given index to a new point in space.
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
nodeIndex the index of the node to be moved rNewLocation the new target location of the node
Implemented in AbstractOffLatticeCellPopulation< DIM >, AbstractOnLatticeCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
| void AbstractCellPopulation< DIM >::SetOutputCellAges | ( | bool | outputCellAges | ) | [inline] |
Set mOutputCellAges.
- Parameters:
-
outputCellAges the new value of mOutputCellAges
Definition at line 819 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellAges.
| void AbstractCellPopulation< DIM >::SetOutputCellAncestors | ( | bool | outputCellAncestors | ) | [inline] |
Set mOutputCellAncestors.
- Parameters:
-
outputCellAncestors the new value of mOutputCellAncestors
Definition at line 795 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellAncestors.
| void AbstractCellPopulation< DIM >::SetOutputCellCyclePhases | ( | bool | outputCellCyclePhases | ) | [inline] |
Set mOutputCellCyclePhases.
- Parameters:
-
outputCellCyclePhases the new value of mOutputCellCyclePhases
Definition at line 813 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellCyclePhases.
| void AbstractCellPopulation< DIM >::SetOutputCellIdData | ( | bool | outputCellIdData | ) | [inline] |
Set mOutputCellIdData.
- Parameters:
-
outputCellIdData the new value of mOutputCellIdData
Definition at line 783 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellIdData.
| void AbstractCellPopulation< DIM >::SetOutputCellMutationStates | ( | bool | outputCellMutationStates | ) | [inline] |
Set mOutputCellMutationStates.
- Parameters:
-
outputCellMutationStates the new value of mOutputCellMutationStates
Definition at line 789 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellMutationStates.
| void AbstractCellPopulation< DIM >::SetOutputCellProliferativeTypes | ( | bool | outputCellProliferativeTypes | ) | [inline] |
Set mOutputCellProliferativeTypes.
- Parameters:
-
outputCellProliferativeTypes the new value of mOutputCellProliferativeTypes
Definition at line 801 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes.
| void AbstractCellPopulation< DIM >::SetOutputCellVariables | ( | bool | outputCellVariables | ) | [inline] |
Set mOutputCellVariables.
- Parameters:
-
outputCellVariables the new value of mOutputCellVariables
Definition at line 807 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellVariables.
| void AbstractCellPopulation< DIM >::SetOutputCellVolumes | ( | bool | outputCellVolumes | ) | [inline] |
Set mOutputCellVolumes.
- Parameters:
-
outputCellVolumes the new value of mOutputCellVolumes
Definition at line 825 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::mOutputCellVolumes.
| virtual void AbstractCellPopulation< DIM >::Update | ( | bool | hasHadBirthsOrDeaths = true |
) | [pure virtual] |
Remove the Nodes (for cell-centre) or VertexElements (for cell-vertex) which have been marked as deleted and update the correspondence with Cells.
- Parameters:
-
hasHadBirthsOrDeaths - a bool saying whether cell population has had Births Or Deaths
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
| virtual void AbstractCellPopulation< DIM >::Validate | ( | ) | [protected, pure virtual] |
Check consistency of our internal data structures.
As this method is pure virtual, it must be overridden in subclasses.
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, MeshBasedCellPopulationWithGhostNodes< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
| void AbstractCellPopulation< DIM >::WriteCellIdDataToFile | ( | ) | [inline] |
Write cell ID data to mpCellIdFile.
Definition at line 650 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::Begin(), AbstractCellPopulation< DIM >::End(), AbstractCellPopulation< DIM >::GetLocationOfCellCentre(), SimulationTime::GetTime(), SimulationTime::Instance(), AbstractCellPopulation< DIM >::mCellLocationMap, and AbstractCellPopulation< DIM >::mpCellIdFile.
Referenced by AbstractCellPopulation< DIM >::WriteResultsToFiles().
| void AbstractCellPopulation< DIM >::WriteCellResultsToFiles | ( | std::vector< unsigned > & | rCellProliferativeTypeCounter, | |
| std::vector< unsigned > & | rCellCyclePhaseCounter | |||
| ) | [inline] |
Write the current state of each cell to output files.
- Parameters:
-
rCellProliferativeTypeCounter cell type counter rCellCyclePhaseCounter cell cycle phase counter
Definition at line 510 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::GetCellMutationStateCount(), AbstractCellPopulation< DIM >::mCellCyclePhaseCount, AbstractCellPopulation< DIM >::mCellProliferativeTypeCount, AbstractCellPopulation< DIM >::mOutputCellAges, AbstractCellPopulation< DIM >::mOutputCellAncestors, AbstractCellPopulation< DIM >::mOutputCellCyclePhases, AbstractCellPopulation< DIM >::mOutputCellMutationStates, AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes, AbstractCellPopulation< DIM >::mOutputCellVariables, AbstractCellPopulation< DIM >::mpCellAgesFile, AbstractCellPopulation< DIM >::mpCellCyclePhasesFile, AbstractCellPopulation< DIM >::mpCellMutationStatesFile, AbstractCellPopulation< DIM >::mpCellProliferativeTypesFile, AbstractCellPopulation< DIM >::mpCellVariablesFile, AbstractCellPopulation< DIM >::mpVizCellAncestorsFile, AbstractCellPopulation< DIM >::mpVizCellProliferativePhasesFile, and AbstractCellPopulation< DIM >::mpVizCellProliferativeTypesFile.
Referenced by VertexBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), PottsBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), CaBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), and AbstractCentreBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles().
| virtual void AbstractCellPopulation< DIM >::WriteCellVolumeResultsToFile | ( | ) | [pure virtual] |
Write the current volume of each cell to file. As this method is pure virtual, it must be overridden in subclasses.
Implemented in CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Referenced by AbstractCellPopulation< DIM >::WriteResultsToFiles().
| void AbstractCellPopulation< DIM >::WriteResultsToFiles | ( | ) | [inline, virtual] |
Write results from the current cell population state to output files.
Reimplemented in MeshBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Definition at line 596 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), SimulationTime::GetTime(), SimulationTime::Instance(), AbstractCellPopulation< DIM >::mOutputCellAges, AbstractCellPopulation< DIM >::mOutputCellAncestors, AbstractCellPopulation< DIM >::mOutputCellCyclePhases, AbstractCellPopulation< DIM >::mOutputCellIdData, AbstractCellPopulation< DIM >::mOutputCellMutationStates, AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes, AbstractCellPopulation< DIM >::mOutputCellVariables, AbstractCellPopulation< DIM >::mOutputCellVolumes, AbstractCellPopulation< DIM >::mpCellAgesFile, AbstractCellPopulation< DIM >::mpCellCyclePhasesFile, AbstractCellPopulation< DIM >::mpCellMutationStatesFile, AbstractCellPopulation< DIM >::mpCellProliferativeTypesFile, AbstractCellPopulation< DIM >::mpCellVariablesFile, AbstractCellPopulation< DIM >::mpVizCellAncestorsFile, AbstractCellPopulation< DIM >::mpVizCellProliferativePhasesFile, AbstractCellPopulation< DIM >::mpVizCellProliferativeTypesFile, AbstractCellPopulation< DIM >::WriteCellIdDataToFile(), AbstractCellPopulation< DIM >::WriteCellVolumeResultsToFile(), AbstractCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles(), and AbstractCellPopulation< DIM >::WriteVtkResultsToFile().
| void AbstractCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles | ( | ) | [inline, virtual] |
Write the current time and node results to output files.
Reimplemented in AbstractCentreBasedCellPopulation< DIM >.
Definition at line 570 of file AbstractCellPopulation.cpp.
References AbstractCellPopulation< DIM >::GetNode(), AbstractCellPopulation< DIM >::GetNumNodes(), SimulationTime::GetTime(), SimulationTime::Instance(), AbstractCellPopulation< DIM >::mpVizBoundaryNodesFile, and AbstractCellPopulation< DIM >::mpVizNodesFile.
Referenced by AbstractCellPopulation< DIM >::WriteResultsToFiles().
| virtual void AbstractCellPopulation< DIM >::WriteVtkResultsToFile | ( | ) | [protected, pure virtual] |
Write the current results to mpVtkMetaFile.
As this method is pure virtual, it must be overridden in subclasses.
Implemented in AbstractCentreBasedCellPopulation< DIM >, CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, MeshBasedCellPopulationWithGhostNodes< DIM >, NodeBasedCellPopulation< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Referenced by AbstractCellPopulation< DIM >::WriteResultsToFiles().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented in AbstractCentreBasedCellPopulation< DIM >, AbstractOffLatticeCellPopulation< DIM >, AbstractOnLatticeCellPopulation< DIM >, CaBasedCellPopulation< DIM >, MeshBasedCellPopulation< DIM >, MeshBasedCellPopulationWithGhostNodes< DIM >, NodeBasedCellPopulation< DIM >, NodeBasedCellPopulationWithBuskeUpdate< DIM >, PottsBasedCellPopulation< DIM >, VertexBasedCellPopulation< DIM >, MeshBasedCellPopulation< 1 >, and MeshBasedCellPopulation< 2 >.
Definition at line 70 of file AbstractCellPopulation.hpp.
Member Data Documentation
std::vector<unsigned> AbstractCellPopulation< DIM >::mCellCyclePhaseCount [protected] |
Current cell cycle phase counts
Definition at line 111 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::AbstractCellPopulation(), VertexBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), PottsBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), CaBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCentreBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCellPopulation< DIM >::rGetCellCyclePhaseCount(), AbstractCellPopulation< SPACE_DIM >::serialize(), and AbstractCellPopulation< DIM >::WriteCellResultsToFiles().
std::map<Cell*, unsigned> AbstractCellPopulation< DIM >::mCellLocationMap [protected] |
Map cells to location (node or VertexElement) indices
Definition at line 105 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::AbstractCellPopulation(), VertexBasedCellPopulation< DIM >::AddCell(), PottsBasedCellPopulation< DIM >::AddCell(), MeshBasedCellPopulationWithGhostNodes< DIM >::AddCell(), CaBasedCellPopulation< DIM >::AddCell(), AbstractCentreBasedCellPopulation< DIM >::AddCell(), MeshBasedCellPopulation< DIM >::CheckCellPointers(), VertexBasedCellPopulation< DIM >::GetElementCorrespondingToCell(), PottsBasedCellPopulation< DIM >::GetElementCorrespondingToCell(), AbstractCellPopulation< DIM >::GetLocationIndexUsingCell(), VertexBasedCellPopulation< DIM >::GetLocationOfCellCentre(), PottsBasedCellPopulation< DIM >::GetLocationOfCellCentre(), CaBasedCellPopulation< DIM >::GetLocationOfCellCentre(), AbstractCentreBasedCellPopulation< DIM >::GetNodeCorrespondingToCell(), CaBasedCellPopulation< DIM >::MoveCell(), VertexBasedCellPopulation< DIM >::RemoveDeadCells(), PottsBasedCellPopulation< DIM >::RemoveDeadCells(), NodeBasedCellPopulation< DIM >::RemoveDeadCells(), MeshBasedCellPopulation< DIM >::RemoveDeadCells(), CaBasedCellPopulation< DIM >::RemoveDeadCells(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetCellAncestorsToLocationIndices(), VertexBasedCellPopulation< DIM >::Update(), NodeBasedCellPopulation< DIM >::Update(), MeshBasedCellPopulation< DIM >::Update(), NodeBasedCellPopulationWithBuskeUpdate< DIM >::UpdateNodeLocations(), AbstractCentreBasedCellPopulation< DIM >::UpdateNodeLocations(), NodeBasedCellPopulation< DIM >::Validate(), MeshBasedCellPopulationWithGhostNodes< DIM >::Validate(), CaBasedCellPopulation< DIM >::Validate(), and AbstractCellPopulation< DIM >::WriteCellIdDataToFile().
std::vector<unsigned> AbstractCellPopulation< DIM >::mCellProliferativeTypeCount [protected] |
Current cell type counts
Definition at line 108 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::AbstractCellPopulation(), VertexBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), PottsBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), CaBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCentreBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCellPopulation< DIM >::rGetCellProliferativeTypeCount(), AbstractCellPopulation< SPACE_DIM >::serialize(), and AbstractCellPopulation< DIM >::WriteCellResultsToFiles().
std::list<CellPtr> AbstractCellPopulation< DIM >::mCells [protected] |
List of cells
Definition at line 99 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::AbstractCellPopulation(), VertexBasedCellPopulation< DIM >::AddCell(), PottsBasedCellPopulation< DIM >::AddCell(), CaBasedCellPopulation< DIM >::AddCell(), AbstractCentreBasedCellPopulation< DIM >::AddCell(), AbstractCellPopulation< DIM >::Begin(), CaBasedCellPopulation< DIM >::CaBasedCellPopulation(), MeshBasedCellPopulation< DIM >::CheckCellPointers(), AbstractCellPopulation< DIM >::End(), MeshBasedCellPopulation< DIM >::MeshBasedCellPopulation(), VertexBasedCellPopulation< DIM >::RemoveDeadCells(), PottsBasedCellPopulation< DIM >::RemoveDeadCells(), NodeBasedCellPopulation< DIM >::RemoveDeadCells(), MeshBasedCellPopulation< DIM >::RemoveDeadCells(), CaBasedCellPopulation< DIM >::RemoveDeadCells(), AbstractCellPopulation< DIM >::rGetCells(), AbstractCellPopulation< SPACE_DIM >::serialize(), VertexBasedCellPopulation< DIM >::Update(), NodeBasedCellPopulation< DIM >::Update(), MeshBasedCellPopulation< DIM >::Update(), VertexBasedCellPopulation< DIM >::WriteResultsToFiles(), and PottsBasedCellPopulation< DIM >::WriteResultsToFiles().
c_vector<double, DIM> AbstractCellPopulation< DIM >::mCentroid [protected] |
Population centroid
Definition at line 114 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::GetCentroidOfCellPopulation().
std::string AbstractCellPopulation< DIM >::mDirPath [protected] |
A cache of where the results are going (used for VTK writer).
Definition at line 153 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CreateOutputFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), PottsBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
std::map<unsigned, CellPtr> AbstractCellPopulation< DIM >::mLocationCellMap [protected] |
Map location (node or VertexElement) indices back to cells
Definition at line 102 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::AbstractCellPopulation(), VertexBasedCellPopulation< DIM >::AddCell(), PottsBasedCellPopulation< DIM >::AddCell(), CaBasedCellPopulation< DIM >::AddCell(), AbstractCentreBasedCellPopulation< DIM >::AddCell(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCentreBasedCellPopulation< DIM >::GenerateCellResultsAndWriteToFiles(), AbstractCellPopulation< DIM >::GetCellUsingLocationIndex(), CaBasedCellPopulation< DIM >::MoveCell(), NodeBasedCellPopulation< DIM >::RemoveDeadCells(), MeshBasedCellPopulation< DIM >::RemoveDeadCells(), CaBasedCellPopulation< DIM >::RemoveDeadCells(), AbstractCellPopulation< SPACE_DIM >::serialize(), VertexBasedCellPopulation< DIM >::Update(), NodeBasedCellPopulation< DIM >::Update(), MeshBasedCellPopulation< DIM >::Update(), MeshBasedCellPopulation< DIM >::WriteCellPopulationVolumeResultsToFile(), VertexBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), PottsBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), MeshBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), VertexBasedCellPopulation< DIM >::WriteResultsToFiles(), PottsBasedCellPopulation< DIM >::WriteResultsToFiles(), MeshBasedCellPopulation< DIM >::WriteResultsToFiles(), AbstractCentreBasedCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), PottsBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
bool AbstractCellPopulation< DIM >::mOutputCellAges [protected] |
Whether to write the cell ages to a file.
Definition at line 180 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GetOutputCellAges(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellAges(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), AbstractCellPopulation< DIM >::WriteResultsToFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
bool AbstractCellPopulation< DIM >::mOutputCellAncestors [protected] |
Whether to output the ancestor of each cell to a visualizer file.
Definition at line 168 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GetOutputCellAncestors(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellAncestors(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), AbstractCellPopulation< DIM >::WriteResultsToFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
bool AbstractCellPopulation< DIM >::mOutputCellCyclePhases [protected] |
Whether to write the cell cycle phases to a file.
Definition at line 177 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GetOutputCellCyclePhases(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< DIM >::rGetCellCyclePhaseCount(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellCyclePhases(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), AbstractCellPopulation< DIM >::WriteResultsToFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
bool AbstractCellPopulation< DIM >::mOutputCellIdData [protected] |
Whether to write cell ID data to file.
Definition at line 162 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GetOutputCellIdData(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellIdData(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
bool AbstractCellPopulation< DIM >::mOutputCellMutationStates [protected] |
Whether to count the number of each cell mutation state and output to file.
Definition at line 165 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GetCellMutationStateCount(), AbstractCellPopulation< DIM >::GetOutputCellMutationStates(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellMutationStates(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), AbstractCellPopulation< DIM >::WriteResultsToFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), PottsBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
bool AbstractCellPopulation< DIM >::mOutputCellProliferativeTypes [protected] |
Whether to count the number of each cell type and output to file.
Definition at line 171 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GetOutputCellProliferativeTypes(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< DIM >::rGetCellProliferativeTypeCount(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellProliferativeTypes(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), AbstractCellPopulation< DIM >::WriteResultsToFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
bool AbstractCellPopulation< DIM >::mOutputCellVariables [protected] |
Whether to write the cell variables to a file.
Definition at line 174 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GetOutputCellVariables(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellVariables(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
bool AbstractCellPopulation< DIM >::mOutputCellVolumes [protected] |
Whether to write the cell volumes (in 3D) or areas (in 2D) to a file.
Definition at line 183 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GetOutputCellVolumes(), AbstractCellPopulation< DIM >::OutputCellPopulationParameters(), AbstractCellPopulation< SPACE_DIM >::serialize(), AbstractCellPopulation< DIM >::SetOutputCellVolumes(), MeshBasedCellPopulation< DIM >::TessellateIfNeeded(), AbstractCellPopulation< DIM >::WriteResultsToFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
out_stream AbstractCellPopulation< DIM >::mpCellAgesFile [protected] |
Results file for cell ages
Definition at line 141 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpCellCyclePhasesFile [protected] |
Results file for cell cycle phases
Definition at line 135 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpCellIdFile [protected] |
Results file for logged cell data.
Definition at line 144 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), and AbstractCellPopulation< DIM >::WriteCellIdDataToFile().
out_stream AbstractCellPopulation< DIM >::mpCellMutationStatesFile [protected] |
Results file for cell mutation states
Definition at line 126 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpCellProliferativeTypesFile [protected] |
Results file for cell types
Definition at line 132 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
boost::shared_ptr<CellPropertyRegistry> AbstractCellPopulation< DIM >::mpCellPropertyRegistry [protected] |
Cell property registry.
Definition at line 159 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::AbstractCellPopulation(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GetCellMutationStateCount(), AbstractCellPopulation< DIM >::GetCellPropertyRegistry(), and AbstractCellPopulation< SPACE_DIM >::serialize().
out_stream AbstractCellPopulation< DIM >::mpCellVariablesFile [protected] |
Results file for cell variables
Definition at line 138 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpCellVolumesFile [protected] |
Results file for cell volume (in 3D) or area (in 2D) data.
Definition at line 147 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), VertexBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), PottsBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), NodeBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), MeshBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile(), and CaBasedCellPopulation< DIM >::WriteCellVolumeResultsToFile().
out_stream AbstractCellPopulation< DIM >::mpVizBoundaryNodesFile [protected] |
Results file for boundary nodes.
Definition at line 150 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCentreBasedCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles(), and AbstractCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpVizCellAncestorsFile [protected] |
Results file for cell ancestors
Definition at line 129 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpVizCellProliferativePhasesFile [protected] |
Results file for cell visualization
Definition at line 123 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpVizCellProliferativeTypesFile [protected] |
Results file for cell visualization
Definition at line 120 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), CaBasedCellPopulation< DIM >::GenerateCellResults(), AbstractCentreBasedCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::GenerateCellResults(), AbstractCellPopulation< DIM >::WriteCellResultsToFiles(), and AbstractCellPopulation< DIM >::WriteResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpVizNodesFile [protected] |
Results file for node visualization
Definition at line 117 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), AbstractCentreBasedCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles(), and AbstractCellPopulation< DIM >::WriteTimeAndNodeResultsToFiles().
out_stream AbstractCellPopulation< DIM >::mpVtkMetaFile [protected] |
Meta results file for VTK.
Definition at line 156 of file AbstractCellPopulation.hpp.
Referenced by AbstractCellPopulation< DIM >::CloseOutputFiles(), AbstractCellPopulation< DIM >::CreateOutputFiles(), VertexBasedCellPopulation< DIM >::WriteVtkResultsToFile(), PottsBasedCellPopulation< DIM >::WriteVtkResultsToFile(), NodeBasedCellPopulation< DIM >::WriteVtkResultsToFile(), MeshBasedCellPopulationWithGhostNodes< DIM >::WriteVtkResultsToFile(), and MeshBasedCellPopulation< DIM >::WriteVtkResultsToFile().
The documentation for this class was generated from the following files:
- cell_based/src/population/AbstractCellPopulation.hpp
- cell_based/src/population/AbstractCellPopulation.cpp
 1.6.3
1.6.3