#include <AbstractCvodeCell.hpp>
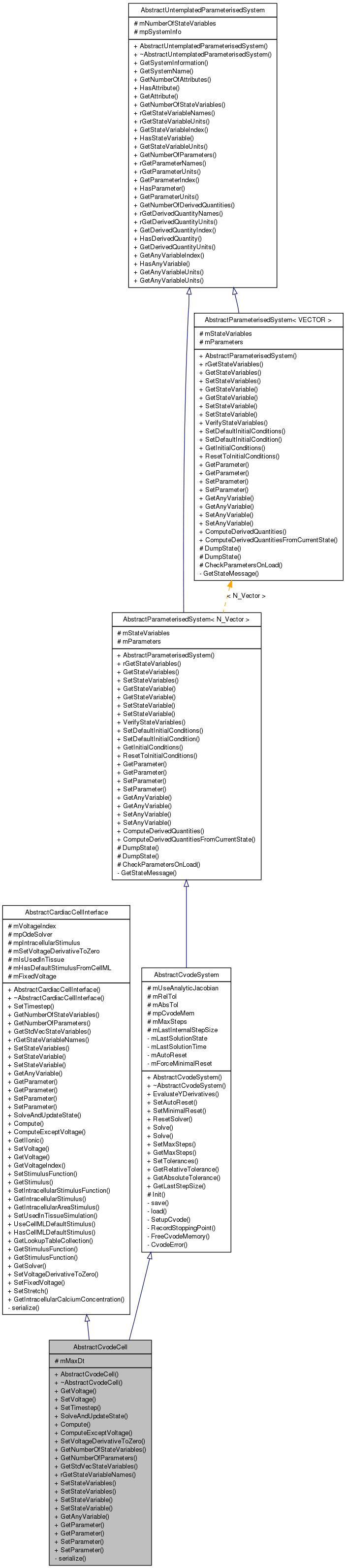
 Inheritance diagram for AbstractCvodeCell:
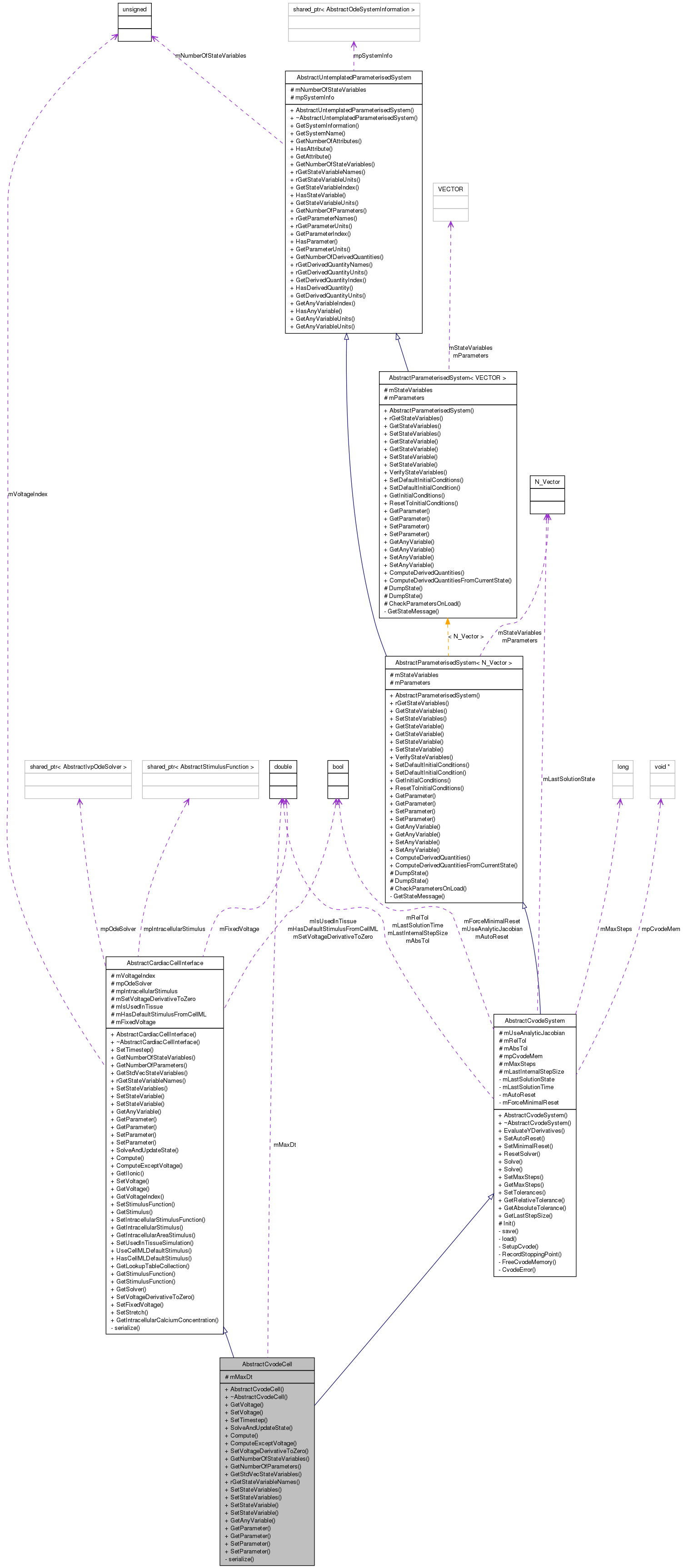
Inheritance diagram for AbstractCvodeCell: Collaboration diagram for AbstractCvodeCell:
Collaboration diagram for AbstractCvodeCell:Public Member Functions | |
| AbstractCvodeCell (boost::shared_ptr< AbstractIvpOdeSolver > pSolver, unsigned numberOfStateVariables, unsigned voltageIndex, boost::shared_ptr< AbstractStimulusFunction > pIntracellularStimulus) | |
| virtual | ~AbstractCvodeCell () |
| double | GetVoltage () |
| void | SetVoltage (double voltage) |
| void | SetTimestep (double maxDt) |
| virtual void | SolveAndUpdateState (double tStart, double tEnd) |
| OdeSolution | Compute (double tStart, double tEnd, double tSamp=0.0) |
| void | ComputeExceptVoltage (double tStart, double tEnd) |
| void | SetVoltageDerivativeToZero (bool clamp=true) |
| unsigned | GetNumberOfStateVariables () const |
| unsigned | GetNumberOfParameters () const |
| std::vector< double > | GetStdVecStateVariables () |
| const std::vector< std::string > & | rGetStateVariableNames () const |
| void | SetStateVariables (const std::vector< double > &rVariables) |
| void | SetStateVariables (const N_Vector &rVariables) |
| void | SetStateVariable (unsigned index, double newValue) |
| void | SetStateVariable (const std::string &rName, double newValue) |
| double | GetAnyVariable (const std::string &rName, double time=0.0) |
| double | GetParameter (const std::string &rParameterName) |
| double | GetParameter (unsigned parameterIndex) |
| void | SetParameter (const std::string &rParameterName, double value) |
| void | SetParameter (unsigned parameterIndex, double value) |
Protected Attributes | |
| double | mMaxDt |
Private Member Functions | |
| template<class Archive > | |
| void | serialize (Archive &archive, const unsigned int version) |
Friends | |
| class | boost::serialization::access |
Detailed Description
A cardiac cell that is designed to be simulated using CVODE. It uses CVODE's vector type natively via AbstractCvodeSystem.
Functionality is similar to that provided by AbstractCardiacCell and AbstractOdeSystem, but not identical. It also includes a direct interface to the CVODE solver, via the Solve methods, since the CvodeAdaptor class may be a bit slower.
Assumes that it will be solving stiff systems, so uses BDF/Newton.
Various methods in this class just call methods on AbstractCvodeSystem, to reduce compiler confusion when working with things in an inheritance tree.
Any single cell work should generally use this class rather than AbstractCardiacCell.
This class may also be faster for certain tissue problems (especially when using a relatively large PDE timestep (~0.1ms)). BUT it will come with a memory overhead as every node has to carry a CVODE solver object as it stores the internal state of the solver, not just the state variables as ForwardEuler solver does. So may not be appropriate for very large meshes.
Definition at line 78 of file AbstractCvodeCell.hpp.
Constructor & Destructor Documentation
| AbstractCvodeCell::AbstractCvodeCell | ( | boost::shared_ptr< AbstractIvpOdeSolver > | pSolver, |
| unsigned | numberOfStateVariables, | ||
| unsigned | voltageIndex, | ||
| boost::shared_ptr< AbstractStimulusFunction > | pIntracellularStimulus | ||
| ) |
Create a new cardiac cell.
- Note:
- subclasses must call Init() in their constructor after setting mpSystemInfo.
- Parameters:
-
pSolver not used for these cells (they're always solved with CVODE); may be empty numberOfStateVariables the size of the ODE system modelling this cell voltageIndex the index of the transmembrane potential within the vector of state variables pIntracellularStimulus the intracellular stimulus current
Definition at line 48 of file AbstractCvodeCell.cpp.
| AbstractCvodeCell::~AbstractCvodeCell | ( | ) | [virtual] |
Free the state variables, if they have been set.
Definition at line 59 of file AbstractCvodeCell.cpp.
Member Function Documentation
| OdeSolution AbstractCvodeCell::Compute | ( | double | tStart, |
| double | tEnd, | ||
| double | tSamp = 0.0 |
||
| ) | [virtual] |
Simulates this cell's behaviour between the time interval [tStart, tEnd], and return state variable values.
The maximum time step to use will be taken as mMaxDt. If this is unset it is the same as the sampling interval, which defaults to HeartConfig::Instance()->GetPrintingTimeStep().
- Parameters:
-
tStart beginning of the time interval to simulate tEnd end of the time interval to simulate tSamp sampling interval for returned results (defaults to dt)
Implements AbstractCardiacCellInterface.
Definition at line 92 of file AbstractCvodeCell.cpp.
References DOUBLE_UNSET, HeartConfig::GetPrintingTimeStep(), HeartConfig::Instance(), mMaxDt, SetTimestep(), and AbstractCvodeSystem::Solve().
Simulates this cell's behaviour between the time interval [tStart, tEnd], but does not update the voltage.
- Parameters:
-
tStart beginning of the time interval to simulate tEnd end of the time interval to simulate
Implements AbstractCardiacCellInterface.
Definition at line 106 of file AbstractCvodeCell.cpp.
References GetVoltage(), SetVoltage(), SetVoltageDerivativeToZero(), SolveAndUpdateState(), and AbstractParameterisedSystem< N_Vector >::VerifyStateVariables().
This just calls the method AbstractCvodeSystem::GetAnyVariable
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Parameters:
-
rName variable name time the time at which to evaluate variable (only needed for derived quantities).
- Returns:
- value of the variable at that time
Implements AbstractCardiacCellInterface.
Definition at line 177 of file AbstractCvodeCell.cpp.
Referenced by GetVoltage().
| unsigned AbstractCvodeCell::GetNumberOfParameters | ( | ) | const [virtual] |
This just returns the number of parameters in the cell model.
It is here because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface
- Returns:
- the number of parameters
Implements AbstractCardiacCellInterface.
Definition at line 138 of file AbstractCvodeCell.cpp.
| unsigned AbstractCvodeCell::GetNumberOfStateVariables | ( | ) | const [virtual] |
This just returns the number of state variables in the cell model.
It is here because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface
- Returns:
- the number of state variables
Implements AbstractCardiacCellInterface.
Definition at line 133 of file AbstractCvodeCell.cpp.
Referenced by GetStdVecStateVariables().
| double AbstractCvodeCell::GetParameter | ( | const std::string & | rParameterName | ) | [virtual] |
This just calls AbstractCvodeSystem::GetParameter
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Parameters:
-
rParameterName the name of a parameter to get the value of,
- Returns:
- the parameter's value.
Implements AbstractCardiacCellInterface.
Definition at line 182 of file AbstractCvodeCell.cpp.
Referenced by GetParameter().
This is just here to show there is an alternative to the above!
It just calls the base class method.
- Parameters:
-
parameterIndex the index of the parameter vector entry to return
- Returns:
- the parameter value
Implements AbstractCardiacCellInterface.
Definition at line 187 of file AbstractCvodeCell.cpp.
References GetParameter().
| std::vector< double > AbstractCvodeCell::GetStdVecStateVariables | ( | ) | [virtual] |
This just returns the state variables in the cell model.
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Returns:
- the state variables
Implements AbstractCardiacCellInterface.
Definition at line 143 of file AbstractCvodeCell.cpp.
References CopyToStdVector(), GetNumberOfStateVariables(), and AbstractParameterisedSystem< N_Vector >::rGetStateVariables().
| double AbstractCvodeCell::GetVoltage | ( | ) | [virtual] |
Get the current value of the transmembrane potential, as given in our state variable vector.
Implements AbstractCardiacCellInterface.
Definition at line 63 of file AbstractCvodeCell.cpp.
References GetAnyVariable(), AbstractParameterisedSystem< N_Vector >::mStateVariables, and AbstractCardiacCellInterface::mVoltageIndex.
Referenced by ComputeExceptVoltage().
| const std::vector< std::string > & AbstractCvodeCell::rGetStateVariableNames | ( | ) | const [virtual] |
Just calls AbstractCvodeSystem::rGetStateVariableNames().
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Returns:
- the state variable names in the cell's ODE system.
Implements AbstractCardiacCellInterface.
Definition at line 150 of file AbstractCvodeCell.cpp.
| void AbstractCvodeCell::serialize | ( | Archive & | archive, |
| const unsigned int | version | ||
| ) | [inline, private] |
Archive the member variables.
- Parameters:
-
archive version
Reimplemented from AbstractCardiacCellInterface.
Definition at line 90 of file AbstractCvodeCell.hpp.
References mMaxDt.
| void AbstractCvodeCell::SetParameter | ( | const std::string & | rParameterName, |
| double | value | ||
| ) | [virtual] |
This just calls AbstractCvodeSystem::SetParameter
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Parameters:
-
rParameterName the parameter name to set the value of, value value to set it to.
Implements AbstractCardiacCellInterface.
Definition at line 192 of file AbstractCvodeCell.cpp.
Referenced by SetParameter().
This is just here to show there is an alternative to the above!
It just calls the base class method.
- Parameters:
-
parameterIndex the index of the parameter vector to alter value the value the parameter should take
Implements AbstractCardiacCellInterface.
Definition at line 197 of file AbstractCvodeCell.cpp.
References SetParameter().
This just calls the method AbstractCvodeSystem::SetStateVariable
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Parameters:
-
index index of the state variable to be set newValue new value of the state variable
Implements AbstractCardiacCellInterface.
Definition at line 167 of file AbstractCvodeCell.cpp.
Referenced by SetStateVariable().
| void AbstractCvodeCell::SetStateVariable | ( | const std::string & | rName, |
| double | newValue | ||
| ) | [virtual] |
This just calls the method AbstractCvodeSystem::SetStateVariable
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Parameters:
-
rName name of the state variable to be set newValue new value of the state variable
Implements AbstractCardiacCellInterface.
Definition at line 172 of file AbstractCvodeCell.cpp.
References SetStateVariable().
| void AbstractCvodeCell::SetStateVariables | ( | const std::vector< double > & | rVariables | ) | [virtual] |
This just sets the state variables in the cell model.
It is here (despite being inherited) because we seem to need to specify explicitly which method in the parent classes we intend to implement to take care of the pure definition in AbstractCardiacCellInterface.
- Parameters:
-
rVariables the state variables (to take a copy of).
Implements AbstractCardiacCellInterface.
Definition at line 155 of file AbstractCvodeCell.cpp.
References DeleteVector(), and MakeNVector().
Referenced by SetStateVariables().
| void AbstractCvodeCell::SetStateVariables | ( | const N_Vector & | rVariables | ) |
This is also needed now just to show there is an alternative to the above method!
- Parameters:
-
rVariables the state variables (to take a copy of).
Reimplemented from AbstractParameterisedSystem< N_Vector >.
Definition at line 162 of file AbstractCvodeCell.cpp.
References SetStateVariables().
| void AbstractCvodeCell::SetTimestep | ( | double | maxDt | ) | [virtual] |
Set the maximum timestep to use for simulating this cell.
- Parameters:
-
maxDt the maximum timestep
Implements AbstractCardiacCellInterface.
Definition at line 77 of file AbstractCvodeCell.cpp.
References mMaxDt.
Referenced by Compute(), and SolveAndUpdateState().
| void AbstractCvodeCell::SetVoltage | ( | double | voltage | ) | [virtual] |
Set the transmembrane potential
- Parameters:
-
voltage new value
Implements AbstractCardiacCellInterface.
Definition at line 69 of file AbstractCvodeCell.cpp.
References AbstractParameterisedSystem< N_Vector >::mStateVariables, AbstractCardiacCellInterface::mVoltageIndex, AbstractParameterisedSystem< N_Vector >::SetAnyVariable(), and AbstractCardiacCellInterface::SetFixedVoltage().
Referenced by ComputeExceptVoltage().
| void AbstractCvodeCell::SetVoltageDerivativeToZero | ( | bool | clamp = true | ) | [virtual] |
Set whether to clamp the voltage by setting its derivative to zero. We need to ensure CVODE is re-initialised if this setting changes.
- Parameters:
-
clamp whether to clamp
Reimplemented from AbstractCardiacCellInterface.
Definition at line 124 of file AbstractCvodeCell.cpp.
References AbstractCardiacCellInterface::mSetVoltageDerivativeToZero, and AbstractCvodeSystem::ResetSolver().
Referenced by ComputeExceptVoltage().
Simulate this cell's behaviour between the time interval [tStart, tEnd], updating the internal state variable values.
The maximum time step to use is given by mMaxDt, which defaults to HeartConfig::Instance()->GetPrintingTimeStep() if unset.
- Parameters:
-
tStart beginning of the time interval to simulate tEnd end of the time interval to simulate
Implements AbstractCardiacCellInterface.
Definition at line 83 of file AbstractCvodeCell.cpp.
References DOUBLE_UNSET, HeartConfig::Instance(), mMaxDt, SetTimestep(), and AbstractCvodeSystem::Solve().
Referenced by ComputeExceptVoltage().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented from AbstractCardiacCellInterface.
Definition at line 82 of file AbstractCvodeCell.hpp.
Member Data Documentation
double AbstractCvodeCell::mMaxDt [protected] |
The maximum timestep to use.
Definition at line 100 of file AbstractCvodeCell.hpp.
Referenced by Compute(), serialize(), SetTimestep(), and SolveAndUpdateState().
The documentation for this class was generated from the following files:
- heart/src/odes/AbstractCvodeCell.hpp
- heart/src/odes/AbstractCvodeCell.cpp