AbstractOdeBasedCellCycleModel Class Reference
#include <AbstractOdeBasedCellCycleModel.hpp>
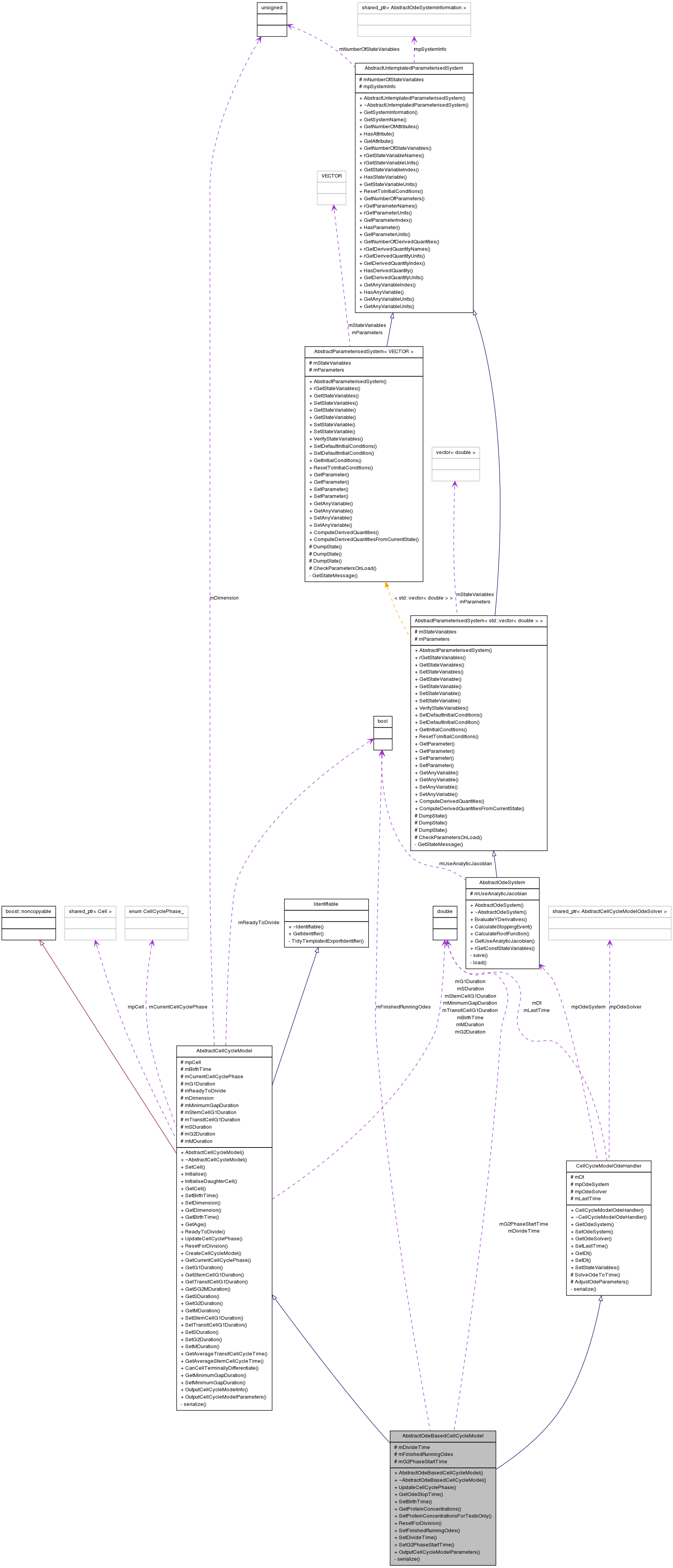
Inherits AbstractCellCycleModel, and CellCycleModelOdeHandler.
Inherited by AbstractWntOdeBasedCellCycleModel, Alarcon2004OxygenBasedCellCycleModel, and TysonNovakCellCycleModel.
Public Member Functions | |
| AbstractOdeBasedCellCycleModel (double lastTime=SimulationTime::Instance()->GetTime(), boost::shared_ptr< AbstractCellCycleModelOdeSolver > pOdeSolver=boost::shared_ptr< AbstractCellCycleModelOdeSolver >()) | |
| virtual | ~AbstractOdeBasedCellCycleModel () |
| virtual void | UpdateCellCyclePhase () |
| double | GetOdeStopTime () |
| void | SetBirthTime (double birthTime) |
| std::vector< double > | GetProteinConcentrations () const |
| void | SetProteinConcentrationsForTestsOnly (double lastTime, std::vector< double > proteinConcentrations) |
| virtual void | ResetForDivision () |
| void | SetFinishedRunningOdes (bool finishedRunningOdes) |
| void | SetDivideTime (double divideTime) |
| void | SetG2PhaseStartTime (double g2PhaseStartTime) |
| virtual void | OutputCellCycleModelParameters (out_stream &rParamsFile) |
Protected Attributes | |
| double | mDivideTime |
| bool | mFinishedRunningOdes |
| double | mG2PhaseStartTime |
Private Member Functions | |
| template<class Archive > | |
| void | serialize (Archive &archive, const unsigned int version) |
Friends | |
| class | boost::serialization::access |
Detailed Description
This class contains all the functionality shared by 'ODE-based' cell-cycle models, where the duration of one or more cell cycle phases are evaluated 'on the fly' as the cell ages, according to a system of ordinary differential equations (ODEs) governing (for example) the concentrations of key intracellular proteins. To determine when cell division should occur, one or more stopping conditions for this ODE system may be specified.
This class of cell-cycle models is distinct from 'simple' cell-cycle models, where the duration of each cell cycle phase is determined when the cell-cycle model is created.
Definition at line 61 of file AbstractOdeBasedCellCycleModel.hpp.
Constructor & Destructor Documentation
| AbstractOdeBasedCellCycleModel::AbstractOdeBasedCellCycleModel | ( | double | lastTime = SimulationTime::Instance()->GetTime(), |
|
| boost::shared_ptr< AbstractCellCycleModelOdeSolver > | pOdeSolver = boost::shared_ptr<AbstractCellCycleModelOdeSolver>() | |||
| ) |
Creates an AbstractOdeBasedCellCycleModel, calls SetBirthTime on the AbstractCellCycleModel to make sure that can be set 'back in time' for cells which did not divide at the current time.
- Parameters:
-
lastTime The birth time of the cell / last time model was evaluated (defaults to the current SimulationTime) pOdeSolver An optional pointer to a cell-cycle model ODE solver object (allows the use of different ODE solvers)
Definition at line 41 of file AbstractOdeBasedCellCycleModel.cpp.
References SetBirthTime().
| AbstractOdeBasedCellCycleModel::~AbstractOdeBasedCellCycleModel | ( | ) | [virtual] |
Destructor.
Definition at line 51 of file AbstractOdeBasedCellCycleModel.cpp.
Member Function Documentation
| double AbstractOdeBasedCellCycleModel::GetOdeStopTime | ( | ) |
Get the time at which the ODE stopping event occurred. Only called in those subclasses for which stopping events are defined.
- Returns:
- the time at which the ODE system reached its stopping event
Definition at line 158 of file AbstractOdeBasedCellCycleModel.cpp.
References DOUBLE_UNSET, and CellCycleModelOdeHandler::mpOdeSolver.
Referenced by AbstractWntOdeBasedCellCycleModel::UpdateCellCyclePhase(), and UpdateCellCyclePhase().
| std::vector< double > AbstractOdeBasedCellCycleModel::GetProteinConcentrations | ( | ) | const |
- Returns:
- the protein concentrations at the current time (useful for tests)
NB: Will copy the vector - you can't use this to modify the concentrations.
Definition at line 62 of file AbstractOdeBasedCellCycleModel.cpp.
References CellCycleModelOdeHandler::mpOdeSystem, and AbstractParameterisedSystem< VECTOR >::rGetStateVariables().
Referenced by CellCycleModelProteinConcentrationsWriter< ELEMENT_DIM, SPACE_DIM >::VisitCell().
| void AbstractOdeBasedCellCycleModel::OutputCellCycleModelParameters | ( | out_stream & | rParamsFile | ) | [virtual] |
Outputs cell cycle model parameters to file.
- Parameters:
-
rParamsFile the file stream to which the parameters are output
Implements AbstractCellCycleModel.
Reimplemented in Alarcon2004OxygenBasedCellCycleModel, TysonNovakCellCycleModel, AbstractVanLeeuwen2009WntSwatCellCycleModel, AbstractWntOdeBasedCellCycleModel, StochasticWntCellCycleModel, VanLeeuwen2009WntSwatCellCycleModelHypothesisOne, VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo, and WntCellCycleModel.
Definition at line 183 of file AbstractOdeBasedCellCycleModel.cpp.
| void AbstractOdeBasedCellCycleModel::ResetForDivision | ( | ) | [virtual] |
For a naturally cycling model this does not need to be overridden in the subclasses. But most models should override this function and then call AbstractOdeBasedCellCycleModel::ResetForDivision() from inside their version.
Reimplemented from AbstractCellCycleModel.
Reimplemented in Alarcon2004OxygenBasedCellCycleModel, TysonNovakCellCycleModel, AbstractWntOdeBasedCellCycleModel, and StochasticWntCellCycleModel.
Definition at line 147 of file AbstractOdeBasedCellCycleModel.cpp.
References AbstractCellCycleModel::mBirthTime, mDivideTime, mFinishedRunningOdes, AbstractCellCycleModel::mG1Duration, and CellCycleModelOdeHandler::mLastTime.
| void AbstractOdeBasedCellCycleModel::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Archive the cell-cycle model and member variables.
- Parameters:
-
archive the archive version the current version of this class
Reimplemented from AbstractCellCycleModel.
Reimplemented in Alarcon2004OxygenBasedCellCycleModel, TysonNovakCellCycleModel, AbstractVanLeeuwen2009WntSwatCellCycleModel, AbstractWntOdeBasedCellCycleModel, StochasticWntCellCycleModel, VanLeeuwen2009WntSwatCellCycleModelHypothesisOne, VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo, and WntCellCycleModel.
Definition at line 74 of file AbstractOdeBasedCellCycleModel.hpp.
References mDivideTime, mFinishedRunningOdes, and mG2PhaseStartTime.
| void AbstractOdeBasedCellCycleModel::SetBirthTime | ( | double | birthTime | ) | [virtual] |
This overrides the AbstractCellCycleModel::SetBirthTime(double birthTime) because an ODE based cell-cycle model has more to reset...
- Parameters:
-
birthTime the simulation time when the cell was born
Reimplemented from AbstractCellCycleModel.
Definition at line 55 of file AbstractOdeBasedCellCycleModel.cpp.
References mDivideTime, and CellCycleModelOdeHandler::mLastTime.
Referenced by AbstractOdeBasedCellCycleModel(), WntCellCycleModel::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisOne::CreateCellCycleModel(), StochasticWntCellCycleModel::CreateCellCycleModel(), TysonNovakCellCycleModel::CreateCellCycleModel(), and Alarcon2004OxygenBasedCellCycleModel::CreateCellCycleModel().
| void AbstractOdeBasedCellCycleModel::SetDivideTime | ( | double | divideTime | ) |
Set mDivideTime.
- Parameters:
-
divideTime the new value of mDivideTime
Definition at line 173 of file AbstractOdeBasedCellCycleModel.cpp.
References mDivideTime.
Referenced by WntCellCycleModel::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisOne::CreateCellCycleModel(), StochasticWntCellCycleModel::CreateCellCycleModel(), TysonNovakCellCycleModel::CreateCellCycleModel(), and Alarcon2004OxygenBasedCellCycleModel::CreateCellCycleModel().
| void AbstractOdeBasedCellCycleModel::SetFinishedRunningOdes | ( | bool | finishedRunningOdes | ) |
Set mFinishedRunningOdes. Used in CreateCellCycleModel().
- Parameters:
-
finishedRunningOdes the new value of mFinishedRunningOdes
Definition at line 168 of file AbstractOdeBasedCellCycleModel.cpp.
References mFinishedRunningOdes.
Referenced by WntCellCycleModel::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisOne::CreateCellCycleModel(), StochasticWntCellCycleModel::CreateCellCycleModel(), TysonNovakCellCycleModel::CreateCellCycleModel(), and Alarcon2004OxygenBasedCellCycleModel::CreateCellCycleModel().
| void AbstractOdeBasedCellCycleModel::SetG2PhaseStartTime | ( | double | g2PhaseStartTime | ) |
Set mG2PhaseStartTime. Used in CreateCellCycleModel().
- Parameters:
-
g2PhaseStartTime the new value of mG2PhaseStartTime
Definition at line 178 of file AbstractOdeBasedCellCycleModel.cpp.
References mG2PhaseStartTime.
Referenced by WntCellCycleModel::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisOne::CreateCellCycleModel(), StochasticWntCellCycleModel::CreateCellCycleModel(), TysonNovakCellCycleModel::CreateCellCycleModel(), and Alarcon2004OxygenBasedCellCycleModel::CreateCellCycleModel().
| void AbstractOdeBasedCellCycleModel::SetProteinConcentrationsForTestsOnly | ( | double | lastTime, | |
| std::vector< double > | proteinConcentrations | |||
| ) |
Sets the protein concentrations and time when the model was last evaluated - should only be called by tests
- Parameters:
-
lastTime the SimulationTime at which the protein concentrations apply proteinConcentrations a standard vector of doubles of protein concentrations
Definition at line 68 of file AbstractOdeBasedCellCycleModel.cpp.
References CellCycleModelOdeHandler::mLastTime, CellCycleModelOdeHandler::mpOdeSystem, AbstractParameterisedSystem< VECTOR >::rGetStateVariables(), and AbstractParameterisedSystem< VECTOR >::SetStateVariables().
| void AbstractOdeBasedCellCycleModel::UpdateCellCyclePhase | ( | ) | [virtual] |
Default UpdateCellCyclePhase() method for an ODE-based cell-cycle model. This method calls SolveOdeToTime() for G1 phase and adds time for the other phases.
Can be overridden if they should do something more subtle.
Implements AbstractCellCycleModel.
Reimplemented in AbstractWntOdeBasedCellCycleModel.
Definition at line 76 of file AbstractOdeBasedCellCycleModel.cpp.
References EXCEPTION, AbstractCellCycleModel::GetAge(), AbstractCellCycleModel::GetG2Duration(), AbstractCellCycleModel::GetMDuration(), AbstractUntemplatedParameterisedSystem::GetNumberOfStateVariables(), GetOdeStopTime(), AbstractCellCycleModel::GetSDuration(), SimulationTime::GetTime(), SimulationTime::Instance(), AbstractCellCycleModel::mBirthTime, AbstractCellCycleModel::mCurrentCellCyclePhase, mDivideTime, mFinishedRunningOdes, AbstractCellCycleModel::mG1Duration, mG2PhaseStartTime, CellCycleModelOdeHandler::mLastTime, CellCycleModelOdeHandler::mpOdeSystem, AbstractParameterisedSystem< VECTOR >::rGetStateVariables(), and CellCycleModelOdeHandler::SolveOdeToTime().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented from AbstractCellCycleModel.
Reimplemented in Alarcon2004OxygenBasedCellCycleModel, TysonNovakCellCycleModel, AbstractVanLeeuwen2009WntSwatCellCycleModel, AbstractWntOdeBasedCellCycleModel, StochasticWntCellCycleModel, VanLeeuwen2009WntSwatCellCycleModelHypothesisOne, VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo, and WntCellCycleModel.
Definition at line 66 of file AbstractOdeBasedCellCycleModel.hpp.
Member Data Documentation
double AbstractOdeBasedCellCycleModel::mDivideTime [protected] |
The time at which the cell should divide - Set this to DBL_MAX in constructor.
Definition at line 86 of file AbstractOdeBasedCellCycleModel.hpp.
Referenced by WntCellCycleModel::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisOne::CreateCellCycleModel(), StochasticWntCellCycleModel::CreateCellCycleModel(), TysonNovakCellCycleModel::CreateCellCycleModel(), Alarcon2004OxygenBasedCellCycleModel::CreateCellCycleModel(), ResetForDivision(), serialize(), SetBirthTime(), SetDivideTime(), and UpdateCellCyclePhase().
Whether the cell-cycle model is currently in a delay (not solving ODEs).
Definition at line 89 of file AbstractOdeBasedCellCycleModel.hpp.
Referenced by WntCellCycleModel::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisOne::CreateCellCycleModel(), StochasticWntCellCycleModel::CreateCellCycleModel(), TysonNovakCellCycleModel::CreateCellCycleModel(), Alarcon2004OxygenBasedCellCycleModel::CreateCellCycleModel(), ResetForDivision(), serialize(), SetFinishedRunningOdes(), and UpdateCellCyclePhase().
The start time for the G2 phase.
Definition at line 92 of file AbstractOdeBasedCellCycleModel.hpp.
Referenced by WntCellCycleModel::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisTwo::CreateCellCycleModel(), VanLeeuwen2009WntSwatCellCycleModelHypothesisOne::CreateCellCycleModel(), StochasticWntCellCycleModel::CreateCellCycleModel(), TysonNovakCellCycleModel::CreateCellCycleModel(), Alarcon2004OxygenBasedCellCycleModel::CreateCellCycleModel(), serialize(), SetG2PhaseStartTime(), and UpdateCellCyclePhase().
The documentation for this class was generated from the following files:
- cell_based/src/population/cell/cycle/AbstractOdeBasedCellCycleModel.hpp
- cell_based/src/population/cell/cycle/AbstractOdeBasedCellCycleModel.cpp
 1.6.2
1.6.2