Documentation for Release 2024.2
Off Lattice Angiogenesis
This tutorial demonstrates functionality for modelling 3D off-lattice angiogenesis in a corneal micro pocket application, similar to that described in Connor et al. 2015 “An integrated approach to quantitative modelling in angiogenesis research”, doi: 10.1098/rsif.2015.0546. It is a 3D simulation modelling VEGF diffusion and decay from an implanted pellet using finite element methods and lattice-free angiogenesis from a large limbal vessel towards the pellet.
The Test
Start by introducing the necessary header files. The first contain functionality for setting up unit tests, smart pointer tools and output management,
#include <cxxtest/TestSuite.h>
#include "SmartPointers.hpp"
#include "OutputFileHandler.hpp"
#include "FileFinder.hpp"
#include "RandomNumberGenerator.hpp"dimensional analysis,
#include "DimensionalChastePoint.hpp"
#include "UnitCollection.hpp"
#include "Owen11Parameters.hpp"
#include "GenericParameters.hpp"
#include "ParameterCollection.hpp"
#include "BaseUnits.hpp"geometry tools,
#include "MappableGridGenerator.hpp"
#include "Part.hpp"vessel networks,
#include "VesselNode.hpp"
#include "VesselNetwork.hpp"
#include "VesselNetworkGenerator.hpp"flow,
#include "VesselImpedanceCalculator.hpp"
#include "FlowSolver.hpp"
#include "ConstantHaematocritSolver.hpp"
#include "StructuralAdaptationSolver.hpp"
#include "WallShearStressCalculator.hpp"
#include "MechanicalStimulusCalculator.hpp"grids and PDEs,
#include "DiscreteContinuumMesh.hpp"
#include "DiscreteContinuumMeshGenerator.hpp"
#include "VtkMeshWriter.hpp"
#include "FiniteElementSolver.hpp"
#include "DiscreteSource.hpp"
#include "VesselBasedDiscreteSource.hpp"
#include "DiscreteContinuumBoundaryCondition.hpp"
#include "LinearSteadyStateDiffusionReactionPde.hpp"
#include "AbstractCellBasedWithTimingsTestSuite.hpp"angiogenesis and regression,
#include "OffLatticeSproutingRule.hpp"
#include "OffLatticeMigrationRule.hpp"
#include "AngiogenesisSolver.hpp"and classes for managing the simulation.
#include "MicrovesselSolver.hpp"This should appear last.
#include "PetscSetupAndFinalize.hpp"
class TestOffLatticeAngiogenesisLiteratePaper : public AbstractCellBasedWithTimingsTestSuite
{
public:
void Test3dLatticeFree() throw(Exception)
{Set up output file management.
MAKE_PTR_ARGS(OutputFileHandler, p_handler, ("TestOffLatticeAngiogenesisLiteratePaper"));
RandomNumberGenerator::Instance()->Reseed(12345);This component uses explicit dimensions for all quantities, but interfaces with solvers which take
non-dimensional inputs. The BaseUnits singleton takes time, length and mass reference scales to
allow non-dimensionalisation when sending quantities to external solvers and re-dimensionalisation of
results. For our purposes microns for length and hours for time are suitable base units.
units::quantity<unit::length> reference_length(1.0 * unit::microns);
units::quantity<unit::time> reference_time(1.0* unit::hours);
BaseUnits::Instance()->SetReferenceLengthScale(reference_length);
BaseUnits::Instance()->SetReferenceTimeScale(reference_time);Set up the domain representing the cornea. This is a thin hemispherical shell. We assume some symmetry to reduce computational expense.
MappableGridGenerator hemisphere_generator;
units::quantity<unit::length> radius(1400.0 * unit::microns);
units::quantity<unit::length> thickness(100.0 * unit::microns);
unsigned num_divisions_x = 10;
unsigned num_divisions_y = 10;
double azimuth_angle = 1.0 * M_PI;
double polar_angle = 0.5 * M_PI;
boost::shared_ptr<Part<3> > p_domain = hemisphere_generator.GenerateHemisphere(radius/reference_length,
thickness/reference_length,
num_divisions_x,
num_divisions_y,
azimuth_angle,
polar_angle);Set up a vessel network, with divisions roughly every ‘cell length’. Initially it is straight. We will map it onto the hemisphere.
VesselNetworkGenerator<3> network_generator;
units::quantity<unit::length> vessel_length = M_PI * radius;
units::quantity<unit::length> cell_length(40.0 * unit::microns);
boost::shared_ptr<VesselNetwork<3> > p_network = network_generator.GenerateSingleVessel(vessel_length,
DimensionalChastePoint<3>(0.0, 4000.0, 0.0),
unsigned(vessel_length/cell_length) + 1, 0);
p_network->GetNode(0)->GetFlowProperties()->SetIsInputNode(true);
p_network->GetNode(0)->GetFlowProperties()->SetPressure(Owen11Parameters::mpInletPressure->GetValue("User"));
p_network->GetNode(p_network->GetNumberOfNodes()-1)->GetFlowProperties()->SetIsOutputNode(true);
p_network->GetNode(p_network->GetNumberOfNodes()-1)->GetFlowProperties()->SetPressure(Owen11Parameters::mpOutletPressure->GetValue("User"));
std::vector<boost::shared_ptr<VesselNode<3> > > nodes = p_network->GetNodes();
for(unsigned idx =0; idx<nodes.size(); idx++)
{
double node_azimuth_angle = azimuth_angle * nodes[idx]->rGetLocation()[0]*reference_length/vessel_length;
double node_polar_angle = polar_angle*nodes[idx]->rGetLocation()[1]*reference_length/vessel_length;
double dimless_radius = (radius-0.5*thickness)/reference_length;
DimensionalChastePoint<3>new_position(dimless_radius * std::cos(node_azimuth_angle) * std::sin(node_polar_angle),
dimless_radius * std::cos(node_polar_angle),
dimless_radius * std::sin(node_azimuth_angle) * std::sin(node_polar_angle),
reference_length);
nodes[idx]->SetLocation(new_position);
}The initial domain and vessel network now look as follows:

In the experimental assay a pellet containing VEGF is implanted near the top of the cornea. We model this as a fixed concentration of VEGF in a cuboidal region. First set up the vegf sub domain.
boost::shared_ptr<Part<3> > p_vegf_domain = Part<3> ::Create();
units::quantity<unit::length> pellet_side_length(300.0*unit::microns);
p_vegf_domain->AddCuboid(pellet_side_length, pellet_side_length, 5.0*pellet_side_length, DimensionalChastePoint<3>(-150.0,
900.0,
0.0));
p_vegf_domain->Write(p_handler->GetOutputDirectoryFullPath()+"initial_vegf_domain.vtp");Now make a finite element mesh on the cornea.
DiscreteContinuumMeshGenerator<3> mesh_generator;
mesh_generator.SetDomain(p_domain);
mesh_generator.SetMaxElementArea(100000.0);
mesh_generator.Update();
boost::shared_ptr<DiscreteContinuumMesh<3> > p_mesh = mesh_generator.GetMesh();Set up the vegf pde
boost::shared_ptr<LinearSteadyStateDiffusionReactionPde<3> > p_vegf_pde = LinearSteadyStateDiffusionReactionPde<3>::Create();
p_vegf_pde->SetIsotropicDiffusionConstant(Owen11Parameters::mpVegfDiffusivity->GetValue("User"));
p_vegf_pde->SetContinuumLinearInUTerm(-Owen11Parameters::mpVegfDecayRate->GetValue("User"));
p_vegf_pde->SetMesh(p_mesh);
p_vegf_pde->SetUseRegularGrid(false);
p_vegf_pde->SetReferenceConcentration(1.e-9*unit::mole_per_metre_cubed);Add a boundary condition to fix the VEGF concentration in the vegf subdomain.
boost::shared_ptr<DiscreteContinuumBoundaryCondition<3> > p_vegf_boundary = DiscreteContinuumBoundaryCondition<3>::Create();
p_vegf_boundary->SetType(BoundaryConditionType::IN_PART);
p_vegf_boundary->SetSource(BoundaryConditionSource::PRESCRIBED);
p_vegf_boundary->SetValue(3.e-9*unit::mole_per_metre_cubed);
p_vegf_boundary->SetDomain(p_vegf_domain);Set up the PDE solvers for the vegf problem. Note the scaling of the concentration to nM to avoid numerical precision problems.
boost::shared_ptr<FiniteElementSolver<3> > p_vegf_solver = FiniteElementSolver<3>::Create();
p_vegf_solver->SetPde(p_vegf_pde);
p_vegf_solver->SetLabel("vegf");
p_vegf_solver->SetMesh(p_mesh);
p_vegf_solver->AddBoundaryCondition(p_vegf_boundary);
p_vegf_solver->SetReferenceConcentration(1.e-9*unit::mole_per_metre_cubed);An example of the VEGF solution is shown here:
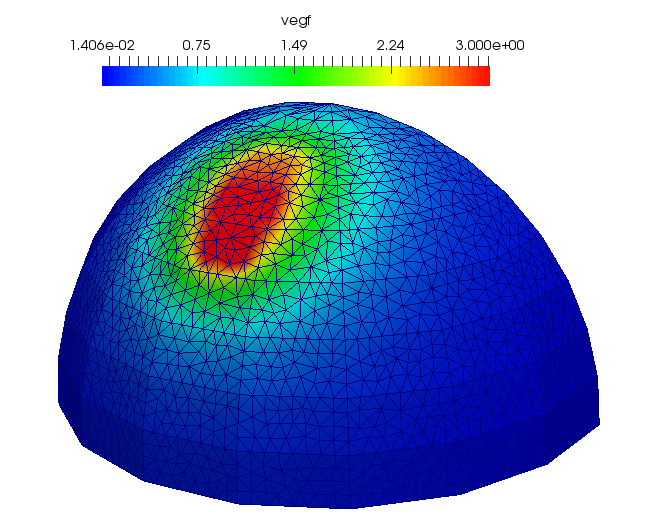
// /*
// * Use the structural adaptation solver to iterate until the flow reaches steady state
// */
// units::quantity<unit::length> vessel_radius(40.0*unit::microns);
// p_network->SetSegmentRadii(vessel_radius);
// units::quantity<unit::dynamic_viscosity> viscosity = Owen11Parameters::mpPlasmaViscosity->GetValue();
// p_network->SetSegmentViscosity(viscosity);
// p_network->Write(p_handler->GetOutputDirectoryFullPath()+"initial_network.vtp");
//
// BaseUnits::Instance()->SetReferenceTimeScale(60.0*unit::seconds);
// SimulationTime::Instance()->SetEndTimeAndNumberOfTimeSteps(30, 1);
// boost::shared_ptr<VesselImpedanceCalculator<3> > p_impedance_calculator = VesselImpedanceCalculator<3>::Create();
// boost::shared_ptr<ConstantHaematocritSolver<3> > p_haematocrit_calculator = ConstantHaematocritSolver<3>::Create();
// p_haematocrit_calculator->SetHaematocrit(0.45);
// boost::shared_ptr<WallShearStressCalculator<3> > p_wss_calculator = WallShearStressCalculator<3>::Create();
// boost::shared_ptr<MechanicalStimulusCalculator<3> > p_mech_stimulus_calculator = MechanicalStimulusCalculator<3>::Create();
//
// StructuralAdaptationSolver<3> structural_adaptation_solver;
// structural_adaptation_solver.SetVesselNetwork(p_network);
// structural_adaptation_solver.SetWriteOutput(true);
// structural_adaptation_solver.SetOutputFileName(p_handler->GetOutputDirectoryFullPath()+"adaptation_data.dat");
// structural_adaptation_solver.SetTolerance(0.0001);
// structural_adaptation_solver.SetMaxIterations(10);
// structural_adaptation_solver.AddPreFlowSolveCalculator(p_impedance_calculator);
// structural_adaptation_solver.AddPostFlowSolveCalculator(p_haematocrit_calculator);
// structural_adaptation_solver.AddPostFlowSolveCalculator(p_wss_calculator);
// structural_adaptation_solver.AddPostFlowSolveCalculator(p_mech_stimulus_calculator);
// structural_adaptation_solver.SetTimeIncrement(0.01 * unit::seconds);
// structural_adaptation_solver.Solve();
Set up an angiogenesis solver and add sprouting and migration rules.
boost::shared_ptr<AngiogenesisSolver<3> > p_angiogenesis_solver = AngiogenesisSolver<3>::Create();
boost::shared_ptr<OffLatticeSproutingRule<3> > p_sprouting_rule = OffLatticeSproutingRule<3>::Create();
p_sprouting_rule->SetSproutingProbability(0.000001* unit::per_second);
boost::shared_ptr<OffLatticeMigrationRule<3> > p_migration_rule = OffLatticeMigrationRule<3>::Create();
p_migration_rule->SetChemotacticStrength(50.0);
units::quantity<unit::velocity> sprout_velocity(20.0*unit::microns/(1.0*unit::hours));
p_migration_rule->SetSproutingVelocity(sprout_velocity);
p_angiogenesis_solver->SetMigrationRule(p_migration_rule);
p_angiogenesis_solver->SetSproutingRule(p_sprouting_rule);
p_sprouting_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_migration_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_angiogenesis_solver->SetVesselNetwork(p_network);
p_angiogenesis_solver->SetBoundingDomain(p_domain);Set up the MicrovesselSolver which coordinates all solves. Note that for sequentially
coupled PDE solves, the solution propagates in the order that the PDE solvers are added to the MicrovesselSolver.
boost::shared_ptr<MicrovesselSolver<3> > p_microvessel_solver = MicrovesselSolver<3>::Create();
p_microvessel_solver->SetVesselNetwork(p_network);
p_microvessel_solver->AddDiscreteContinuumSolver(p_vegf_solver);
p_microvessel_solver->SetOutputFileHandler(p_handler);
p_microvessel_solver->SetOutputFrequency(1);
p_microvessel_solver->SetAngiogenesisSolver(p_angiogenesis_solver);
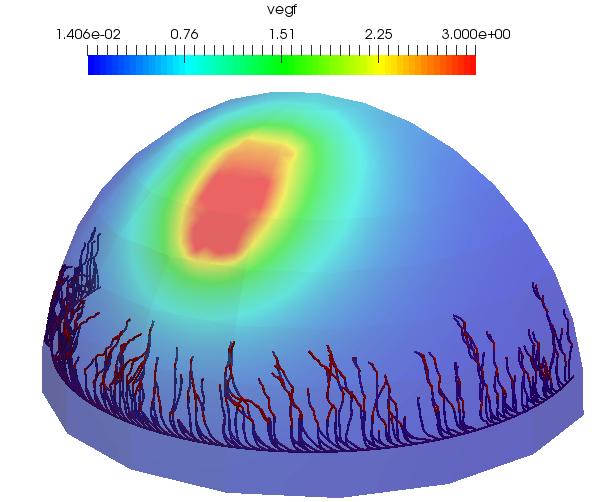
p_microvessel_solver->SetUpdatePdeEachSolve(false);Set the simulation time and run the solver. The result is shown at the top of the tutorial.
SimulationTime::Instance()->SetEndTimeAndNumberOfTimeSteps(100.0, 200);
p_microvessel_solver->Run();
}
};Full code
#include <cxxtest/TestSuite.h>
#include "SmartPointers.hpp"
#include "OutputFileHandler.hpp"
#include "FileFinder.hpp"
#include "RandomNumberGenerator.hpp"
#include "DimensionalChastePoint.hpp"
#include "UnitCollection.hpp"
#include "Owen11Parameters.hpp"
#include "GenericParameters.hpp"
#include "ParameterCollection.hpp"
#include "BaseUnits.hpp"
#include "MappableGridGenerator.hpp"
#include "Part.hpp"
#include "VesselNode.hpp"
#include "VesselNetwork.hpp"
#include "VesselNetworkGenerator.hpp"
#include "VesselImpedanceCalculator.hpp"
#include "FlowSolver.hpp"
#include "ConstantHaematocritSolver.hpp"
#include "StructuralAdaptationSolver.hpp"
#include "WallShearStressCalculator.hpp"
#include "MechanicalStimulusCalculator.hpp"
#include "DiscreteContinuumMesh.hpp"
#include "DiscreteContinuumMeshGenerator.hpp"
#include "VtkMeshWriter.hpp"
#include "FiniteElementSolver.hpp"
#include "DiscreteSource.hpp"
#include "VesselBasedDiscreteSource.hpp"
#include "DiscreteContinuumBoundaryCondition.hpp"
#include "LinearSteadyStateDiffusionReactionPde.hpp"
#include "AbstractCellBasedWithTimingsTestSuite.hpp"
#include "OffLatticeSproutingRule.hpp"
#include "OffLatticeMigrationRule.hpp"
#include "AngiogenesisSolver.hpp"
#include "MicrovesselSolver.hpp"
#include "PetscSetupAndFinalize.hpp"
class TestOffLatticeAngiogenesisLiteratePaper : public AbstractCellBasedWithTimingsTestSuite
{
public:
void Test3dLatticeFree() throw(Exception)
{
MAKE_PTR_ARGS(OutputFileHandler, p_handler, ("TestOffLatticeAngiogenesisLiteratePaper"));
RandomNumberGenerator::Instance()->Reseed(12345);
units::quantity<unit::length> reference_length(1.0 * unit::microns);
units::quantity<unit::time> reference_time(1.0* unit::hours);
BaseUnits::Instance()->SetReferenceLengthScale(reference_length);
BaseUnits::Instance()->SetReferenceTimeScale(reference_time);
MappableGridGenerator hemisphere_generator;
units::quantity<unit::length> radius(1400.0 * unit::microns);
units::quantity<unit::length> thickness(100.0 * unit::microns);
unsigned num_divisions_x = 10;
unsigned num_divisions_y = 10;
double azimuth_angle = 1.0 * M_PI;
double polar_angle = 0.5 * M_PI;
boost::shared_ptr<Part<3> > p_domain = hemisphere_generator.GenerateHemisphere(radius/reference_length,
thickness/reference_length,
num_divisions_x,
num_divisions_y,
azimuth_angle,
polar_angle);
VesselNetworkGenerator<3> network_generator;
units::quantity<unit::length> vessel_length = M_PI * radius;
units::quantity<unit::length> cell_length(40.0 * unit::microns);
boost::shared_ptr<VesselNetwork<3> > p_network = network_generator.GenerateSingleVessel(vessel_length,
DimensionalChastePoint<3>(0.0, 4000.0, 0.0),
unsigned(vessel_length/cell_length) + 1, 0);
p_network->GetNode(0)->GetFlowProperties()->SetIsInputNode(true);
p_network->GetNode(0)->GetFlowProperties()->SetPressure(Owen11Parameters::mpInletPressure->GetValue("User"));
p_network->GetNode(p_network->GetNumberOfNodes()-1)->GetFlowProperties()->SetIsOutputNode(true);
p_network->GetNode(p_network->GetNumberOfNodes()-1)->GetFlowProperties()->SetPressure(Owen11Parameters::mpOutletPressure->GetValue("User"));
std::vector<boost::shared_ptr<VesselNode<3> > > nodes = p_network->GetNodes();
for(unsigned idx =0; idx<nodes.size(); idx++)
{
double node_azimuth_angle = azimuth_angle * nodes[idx]->rGetLocation()[0]*reference_length/vessel_length;
double node_polar_angle = polar_angle*nodes[idx]->rGetLocation()[1]*reference_length/vessel_length;
double dimless_radius = (radius-0.5*thickness)/reference_length;
DimensionalChastePoint<3>new_position(dimless_radius * std::cos(node_azimuth_angle) * std::sin(node_polar_angle),
dimless_radius * std::cos(node_polar_angle),
dimless_radius * std::sin(node_azimuth_angle) * std::sin(node_polar_angle),
reference_length);
nodes[idx]->SetLocation(new_position);
}
boost::shared_ptr<Part<3> > p_vegf_domain = Part<3> ::Create();
units::quantity<unit::length> pellet_side_length(300.0*unit::microns);
p_vegf_domain->AddCuboid(pellet_side_length, pellet_side_length, 5.0*pellet_side_length, DimensionalChastePoint<3>(-150.0,
900.0,
0.0));
p_vegf_domain->Write(p_handler->GetOutputDirectoryFullPath()+"initial_vegf_domain.vtp");
DiscreteContinuumMeshGenerator<3> mesh_generator;
mesh_generator.SetDomain(p_domain);
mesh_generator.SetMaxElementArea(100000.0);
mesh_generator.Update();
boost::shared_ptr<DiscreteContinuumMesh<3> > p_mesh = mesh_generator.GetMesh();
boost::shared_ptr<LinearSteadyStateDiffusionReactionPde<3> > p_vegf_pde = LinearSteadyStateDiffusionReactionPde<3>::Create();
p_vegf_pde->SetIsotropicDiffusionConstant(Owen11Parameters::mpVegfDiffusivity->GetValue("User"));
p_vegf_pde->SetContinuumLinearInUTerm(-Owen11Parameters::mpVegfDecayRate->GetValue("User"));
p_vegf_pde->SetMesh(p_mesh);
p_vegf_pde->SetUseRegularGrid(false);
p_vegf_pde->SetReferenceConcentration(1.e-9*unit::mole_per_metre_cubed);
boost::shared_ptr<DiscreteContinuumBoundaryCondition<3> > p_vegf_boundary = DiscreteContinuumBoundaryCondition<3>::Create();
p_vegf_boundary->SetType(BoundaryConditionType::IN_PART);
p_vegf_boundary->SetSource(BoundaryConditionSource::PRESCRIBED);
p_vegf_boundary->SetValue(3.e-9*unit::mole_per_metre_cubed);
p_vegf_boundary->SetDomain(p_vegf_domain);
boost::shared_ptr<FiniteElementSolver<3> > p_vegf_solver = FiniteElementSolver<3>::Create();
p_vegf_solver->SetPde(p_vegf_pde);
p_vegf_solver->SetLabel("vegf");
p_vegf_solver->SetMesh(p_mesh);
p_vegf_solver->AddBoundaryCondition(p_vegf_boundary);
p_vegf_solver->SetReferenceConcentration(1.e-9*unit::mole_per_metre_cubed);
// /*
// * Use the structural adaptation solver to iterate until the flow reaches steady state
// */
// units::quantity<unit::length> vessel_radius(40.0*unit::microns);
// p_network->SetSegmentRadii(vessel_radius);
// units::quantity<unit::dynamic_viscosity> viscosity = Owen11Parameters::mpPlasmaViscosity->GetValue();
// p_network->SetSegmentViscosity(viscosity);
// p_network->Write(p_handler->GetOutputDirectoryFullPath()+"initial_network.vtp");
//
// BaseUnits::Instance()->SetReferenceTimeScale(60.0*unit::seconds);
// SimulationTime::Instance()->SetEndTimeAndNumberOfTimeSteps(30, 1);
// boost::shared_ptr<VesselImpedanceCalculator<3> > p_impedance_calculator = VesselImpedanceCalculator<3>::Create();
// boost::shared_ptr<ConstantHaematocritSolver<3> > p_haematocrit_calculator = ConstantHaematocritSolver<3>::Create();
// p_haematocrit_calculator->SetHaematocrit(0.45);
// boost::shared_ptr<WallShearStressCalculator<3> > p_wss_calculator = WallShearStressCalculator<3>::Create();
// boost::shared_ptr<MechanicalStimulusCalculator<3> > p_mech_stimulus_calculator = MechanicalStimulusCalculator<3>::Create();
//
// StructuralAdaptationSolver<3> structural_adaptation_solver;
// structural_adaptation_solver.SetVesselNetwork(p_network);
// structural_adaptation_solver.SetWriteOutput(true);
// structural_adaptation_solver.SetOutputFileName(p_handler->GetOutputDirectoryFullPath()+"adaptation_data.dat");
// structural_adaptation_solver.SetTolerance(0.0001);
// structural_adaptation_solver.SetMaxIterations(10);
// structural_adaptation_solver.AddPreFlowSolveCalculator(p_impedance_calculator);
// structural_adaptation_solver.AddPostFlowSolveCalculator(p_haematocrit_calculator);
// structural_adaptation_solver.AddPostFlowSolveCalculator(p_wss_calculator);
// structural_adaptation_solver.AddPostFlowSolveCalculator(p_mech_stimulus_calculator);
// structural_adaptation_solver.SetTimeIncrement(0.01 * unit::seconds);
// structural_adaptation_solver.Solve();
boost::shared_ptr<AngiogenesisSolver<3> > p_angiogenesis_solver = AngiogenesisSolver<3>::Create();
boost::shared_ptr<OffLatticeSproutingRule<3> > p_sprouting_rule = OffLatticeSproutingRule<3>::Create();
p_sprouting_rule->SetSproutingProbability(0.000001* unit::per_second);
boost::shared_ptr<OffLatticeMigrationRule<3> > p_migration_rule = OffLatticeMigrationRule<3>::Create();
p_migration_rule->SetChemotacticStrength(50.0);
units::quantity<unit::velocity> sprout_velocity(20.0*unit::microns/(1.0*unit::hours));
p_migration_rule->SetSproutingVelocity(sprout_velocity);
p_angiogenesis_solver->SetMigrationRule(p_migration_rule);
p_angiogenesis_solver->SetSproutingRule(p_sprouting_rule);
p_sprouting_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_migration_rule->SetDiscreteContinuumSolver(p_vegf_solver);
p_angiogenesis_solver->SetVesselNetwork(p_network);
p_angiogenesis_solver->SetBoundingDomain(p_domain);
boost::shared_ptr<MicrovesselSolver<3> > p_microvessel_solver = MicrovesselSolver<3>::Create();
p_microvessel_solver->SetVesselNetwork(p_network);
p_microvessel_solver->AddDiscreteContinuumSolver(p_vegf_solver);
p_microvessel_solver->SetOutputFileHandler(p_handler);
p_microvessel_solver->SetOutputFrequency(1);
p_microvessel_solver->SetAngiogenesisSolver(p_angiogenesis_solver);
p_microvessel_solver->SetUpdatePdeEachSolve(false);
SimulationTime::Instance()->SetEndTimeAndNumberOfTimeSteps(100.0, 200);
p_microvessel_solver->Run();
}
};