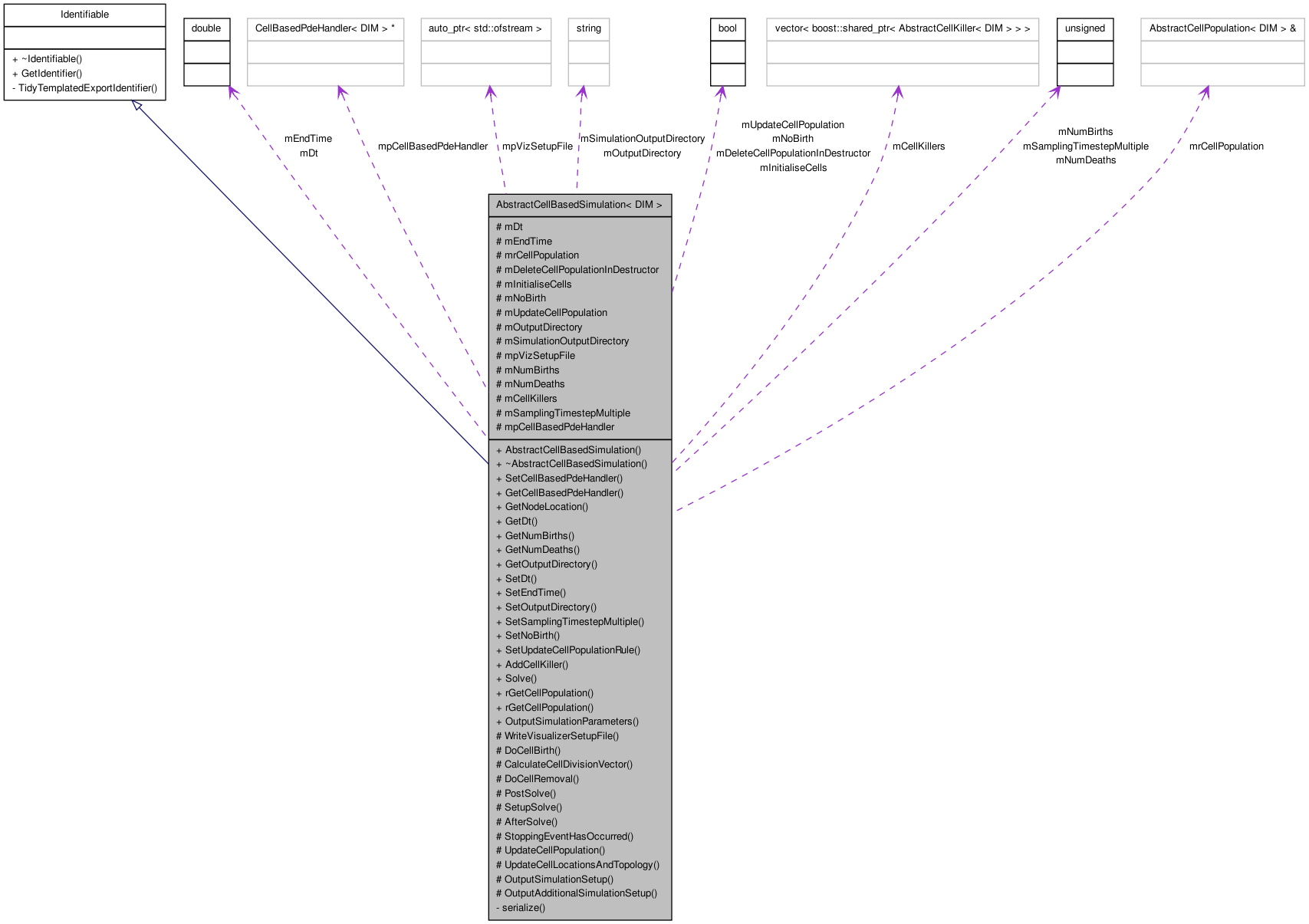
AbstractCellBasedSimulation< DIM > Class Template Reference
#include <AbstractCellBasedSimulation.hpp>
Inherits Identifiable.
Inherited by OffLatticeSimulation< DIM >, OffLatticeSimulation< 1 >, OffLatticeSimulation< 2 >, and OnLatticeSimulation< DIM >.
Detailed Description
template<unsigned DIM>
class AbstractCellBasedSimulation< DIM >
An abstract cell-based simulation class. This class contains common functionality from off lattice and on lattice simulations.
The AbstractCellBasedSimulation is constructed with a CellPopulation, which updates the correspondence between each Cell and its spatial representation and handles cell division (governed by the CellCycleModel associated with each cell). Once constructed, one or more CellKillers may be passed to the AbstractCellBasedSimulation object to specify conditions in which Cells may die,
Subclasses use one or more Force laws or update rules (Which are passed to the child class object) to define the mechanical properties of the CellPopulation.
Definition at line 58 of file AbstractCellBasedSimulation.hpp.
Constructor & Destructor Documentation
| AbstractCellBasedSimulation< DIM >::AbstractCellBasedSimulation | ( | AbstractCellPopulation< DIM > & | rCellPopulation, | |
| bool | deleteCellPopulationInDestructor = false, |
|||
| bool | initialiseCells = true | |||
| ) | [inline] |
Constructor.
- Parameters:
-
rCellPopulation A cell population object deleteCellPopulationInDestructor Whether to delete the cell population on destruction to free up memory (defaults to false) initialiseCells Whether to initialise cells (defaults to true; set to false when loading from an archive)
Definition at line 45 of file AbstractCellBasedSimulation.cpp.
References RandomNumberGenerator::Instance(), AbstractCellBasedSimulation< DIM >::mInitialiseCells, and AbstractCellBasedSimulation< DIM >::mrCellPopulation.
| AbstractCellBasedSimulation< DIM >::~AbstractCellBasedSimulation | ( | ) | [inline, virtual] |
Destructor.
This frees the cell population if it was created by de-serialization.
Definition at line 72 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mDeleteCellPopulationInDestructor, AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler, and AbstractCellBasedSimulation< DIM >::mrCellPopulation.
Member Function Documentation
| void AbstractCellBasedSimulation< DIM >::AddCellKiller | ( | boost::shared_ptr< AbstractCellKiller< DIM > > | pCellKiller | ) | [inline] |
Add a cell killer to be used in this simulation.
- Parameters:
-
pCellKiller pointer to a cell killer
Definition at line 228 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mCellKillers.
| void AbstractCellBasedSimulation< DIM >::AfterSolve | ( | ) | [inline, protected, virtual] |
This method may be overridden in subclasses to do something at the end of each time loop.
Reimplemented in OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, CryptSimulation2d, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Definition at line 266 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler.
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
| virtual c_vector<double, DIM> AbstractCellBasedSimulation< DIM >::CalculateCellDivisionVector | ( | CellPtr | pParentCell | ) | [protected, pure virtual] |
Method for determining how cell division occurs. This method returns a vector which is then passed into the CellPopulation method AddCell().
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
pParentCell the parent cell
- Returns:
- a vector containing information on cell division.
Implemented in OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, CryptSimulation1d, CryptSimulation2d, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Referenced by AbstractCellBasedSimulation< DIM >::DoCellBirth().
| unsigned AbstractCellBasedSimulation< DIM >::DoCellBirth | ( | ) | [inline, protected, virtual] |
During a simulation time step, process any cell divisions that need to occur. If the simulation includes cell birth, causes (almost) all cells that are ready to divide to produce daughter cells.
- Returns:
- the number of births that occurred.
Definition at line 94 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::CalculateCellDivisionVector(), AbstractCellBasedSimulation< DIM >::mNoBirth, and AbstractCellBasedSimulation< DIM >::mrCellPopulation.
Referenced by AbstractCellBasedSimulation< DIM >::UpdateCellPopulation().
| unsigned AbstractCellBasedSimulation< DIM >::DoCellRemoval | ( | ) | [inline, protected] |
During a simulation time step, process any cell sloughing or death
This uses the cell killers to remove cells and associated nodes from the facade class.
- Returns:
- the number of deaths that occurred.
Definition at line 131 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mCellKillers, and AbstractCellBasedSimulation< DIM >::mrCellPopulation.
Referenced by AbstractCellBasedSimulation< DIM >::UpdateCellPopulation().
| CellBasedPdeHandler< DIM > * AbstractCellBasedSimulation< DIM >::GetCellBasedPdeHandler | ( | ) | [inline] |
- Returns:
- mpCellBasedPdeHandler
Definition at line 88 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler.
| double AbstractCellBasedSimulation< DIM >::GetDt | ( | ) | [inline] |
- Returns:
- the timestep of the simulation
Definition at line 159 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mDt.
| std::vector< double > AbstractCellBasedSimulation< DIM >::GetNodeLocation | ( | const unsigned & | rNodeIndex | ) | [inline] |
Get a node's location (ONLY FOR TESTING).
- Parameters:
-
rNodeIndex the node index
- Returns:
- the co-ordinates of this node.
Definition at line 234 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mrCellPopulation.
| unsigned AbstractCellBasedSimulation< DIM >::GetNumBirths | ( | ) | [inline] |
- Returns:
- the number of births that have occurred in the entire simulation (since t=0)
Definition at line 165 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mNumBirths.
| unsigned AbstractCellBasedSimulation< DIM >::GetNumDeaths | ( | ) | [inline] |
- Returns:
- the number of deaths that have occurred in the entire simulation (since t=0).
Definition at line 171 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mNumDeaths.
| std::string AbstractCellBasedSimulation< DIM >::GetOutputDirectory | ( | ) | [inline] |
Get the output directory of the simulation.
Definition at line 191 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mOutputDirectory.
| virtual void AbstractCellBasedSimulation< DIM >::OutputAdditionalSimulationSetup | ( | out_stream & | rParamsFile | ) | [inline, protected, virtual] |
Helper method to output additional simulations parameters and information defined in subclasses to file.
- Parameters:
-
rParamsFile the file stream to which the parameters are output
Reimplemented in OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Definition at line 237 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::OutputSimulationSetup().
| void AbstractCellBasedSimulation< DIM >::OutputSimulationParameters | ( | out_stream & | rParamsFile | ) | [inline, pure virtual] |
Outputs simulation parameters to file
As this method is pure virtual, it must be overridden in subclasses.
- Parameters:
-
rParamsFile the file stream to which the parameters are output
Implemented in OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, CryptSimulation1d, CryptSimulation2d, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Definition at line 483 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mDt, AbstractCellBasedSimulation< DIM >::mEndTime, AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler, and AbstractCellBasedSimulation< DIM >::mSamplingTimestepMultiple.
Referenced by AbstractCellBasedSimulation< DIM >::OutputSimulationSetup().
| void AbstractCellBasedSimulation< DIM >::OutputSimulationSetup | ( | ) | [inline, protected] |
Helper method to output all the simulations parameters and information to file.
Definition at line 437 of file AbstractCellBasedSimulation.cpp.
References Identifiable::GetIdentifier(), AbstractCellBasedSimulation< DIM >::mCellKillers, AbstractCellBasedSimulation< DIM >::mrCellPopulation, AbstractCellBasedSimulation< DIM >::mSimulationOutputDirectory, OutputFileHandler::OpenOutputFile(), AbstractCellBasedSimulation< DIM >::OutputAdditionalSimulationSetup(), AbstractCellBasedSimulation< DIM >::OutputSimulationParameters(), ExecutableSupport::WriteLibraryInfo(), and ExecutableSupport::WriteMachineInfoFile().
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
| void AbstractCellBasedSimulation< DIM >::PostSolve | ( | ) | [inline, protected, virtual] |
A method for subclasses to do something at the end of each timestep
Reimplemented in DeltaNotchOffLatticeSimulation< DIM >, VolumeTrackedOffLatticeSimulation< DIM >, and CryptSimulation2d.
Definition at line 255 of file AbstractCellBasedSimulation.cpp.
References GenericEventHandler< 10, CellBasedEventHandler >::BeginEvent(), GenericEventHandler< 10, CellBasedEventHandler >::EndEvent(), AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler, and AbstractCellBasedSimulation< DIM >::mSamplingTimestepMultiple.
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
| const AbstractCellPopulation< DIM > & AbstractCellBasedSimulation< DIM >::rGetCellPopulation | ( | ) | const [inline] |
- Returns:
- const reference to the cell population (used in archiving).
Definition at line 210 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mrCellPopulation.
| AbstractCellPopulation< DIM > & AbstractCellBasedSimulation< DIM >::rGetCellPopulation | ( | ) | [inline] |
- Returns:
- reference to the cell population.
Definition at line 204 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mrCellPopulation.
| void AbstractCellBasedSimulation< DIM >::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Save or restore the simulation.
- Parameters:
-
archive the archive version the current version of this class
Reimplemented in DeltaNotchOffLatticeSimulation< DIM >, OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, VolumeTrackedOffLatticeSimulation< DIM >, CryptSimulation1d, CryptSimulation2d, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Definition at line 77 of file AbstractCellBasedSimulation.hpp.
References SerializableSingleton< SINGLETON_CLASS >::GetSerializationWrapper(), RandomNumberGenerator::Instance(), SimulationTime::Instance(), AbstractCellBasedSimulation< DIM >::mCellKillers, AbstractCellBasedSimulation< DIM >::mDt, AbstractCellBasedSimulation< DIM >::mEndTime, AbstractCellBasedSimulation< DIM >::mNoBirth, AbstractCellBasedSimulation< DIM >::mNumBirths, AbstractCellBasedSimulation< DIM >::mNumDeaths, AbstractCellBasedSimulation< DIM >::mOutputDirectory, AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler, AbstractCellBasedSimulation< DIM >::mSamplingTimestepMultiple, and AbstractCellBasedSimulation< DIM >::mUpdateCellPopulation.
| void AbstractCellBasedSimulation< DIM >::SetCellBasedPdeHandler | ( | CellBasedPdeHandler< DIM > * | pCellBasedPdeHandler | ) | [inline] |
Set mpCellBasedPdeHandler
- Parameters:
-
pCellBasedPdeHandler pointer to a CellBasedPdeHandler object
Definition at line 82 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler.
| void AbstractCellBasedSimulation< DIM >::SetDt | ( | double | dt | ) | [inline] |
Set the timestep of the simulation.
- Parameters:
-
dt the timestep to use
Definition at line 152 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mDt.
| void AbstractCellBasedSimulation< DIM >::SetEndTime | ( | double | endTime | ) | [inline] |
Set the end time and resets the timestep to be endtime/100.
- Parameters:
-
endTime the end time to use
Definition at line 177 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mEndTime.
| void AbstractCellBasedSimulation< DIM >::SetNoBirth | ( | bool | noBirth | ) | [inline] |
Set the simulation to run with no birth.
- Parameters:
-
noBirth whether to run with no birth
Definition at line 222 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mNoBirth.
| void AbstractCellBasedSimulation< DIM >::SetOutputDirectory | ( | std::string | outputDirectory | ) | [inline] |
Set the output directory of the simulation.
- Parameters:
-
outputDirectory the output directory to use
Definition at line 184 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mOutputDirectory, and AbstractCellBasedSimulation< DIM >::mSimulationOutputDirectory.
| void AbstractCellBasedSimulation< DIM >::SetSamplingTimestepMultiple | ( | unsigned | samplingTimestepMultiple | ) | [inline] |
Set the ratio of the number of actual timesteps to the number of timesteps at which results are written to file. Default value is set to 1 by the constructor.
- Parameters:
-
samplingTimestepMultiple the ratio to use
Definition at line 197 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mSamplingTimestepMultiple.
| void AbstractCellBasedSimulation< DIM >::SetUpdateCellPopulationRule | ( | bool | updateCellPopulation | ) | [inline] |
Set whether to update the topology of the cell population at each time step.
- Parameters:
-
updateCellPopulation whether to update the cell population each time step
Definition at line 216 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mUpdateCellPopulation.
| void AbstractCellBasedSimulation< DIM >::SetupSolve | ( | ) | [inline, protected, virtual] |
A method for subclasses to do something at before the start of the time loop.
Reimplemented in OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, CryptSimulation2d, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Definition at line 245 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler, AbstractCellBasedSimulation< DIM >::mpVizSetupFile, and AbstractCellBasedSimulation< DIM >::mSimulationOutputDirectory.
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
| void AbstractCellBasedSimulation< DIM >::Solve | ( | ) | [inline] |
Main solve method.
This method sets up the simulation time, creates output files, and initialises the cell population. It then iterates through a time loop. At each time step, first any cell death or birth is implemented, then the cell population topology is updated, then the forces are recalculated and the cell population evolved according to whatever force laws are present in the simulation, and finally the results for that time step are output to file. At the end of the time loop, the method closes any output files.
Definition at line 275 of file AbstractCellBasedSimulation.cpp.
References AbstractCellBasedSimulation< DIM >::AfterSolve(), GenericEventHandler< 10, CellBasedEventHandler >::BeginEvent(), GenericEventHandler< 10, CellBasedEventHandler >::EndEvent(), EXCEPTION, SimulationTime::GetTime(), SimulationTime::GetTimeStepsElapsed(), SimulationTime::IncrementTimeOneStep(), SimulationTime::Instance(), SimulationTime::IsEndTimeAndNumberOfTimeStepsSetUp(), SimulationTime::IsFinished(), AbstractCellBasedSimulation< DIM >::mDt, AbstractCellBasedSimulation< DIM >::mEndTime, AbstractCellBasedSimulation< DIM >::mOutputDirectory, AbstractCellBasedSimulation< DIM >::mpVizSetupFile, AbstractCellBasedSimulation< DIM >::mrCellPopulation, AbstractCellBasedSimulation< DIM >::mSamplingTimestepMultiple, AbstractCellBasedSimulation< DIM >::mSimulationOutputDirectory, OutputFileHandler::OpenOutputFile(), AbstractCellBasedSimulation< DIM >::OutputSimulationSetup(), AbstractCellBasedSimulation< DIM >::PostSolve(), SimulationTime::ResetEndTimeAndNumberOfTimeSteps(), SimulationTime::SetEndTimeAndNumberOfTimeSteps(), AbstractCellBasedSimulation< DIM >::SetupSolve(), AbstractCellBasedSimulation< DIM >::StoppingEventHasOccurred(), AbstractCellBasedSimulation< DIM >::UpdateCellLocationsAndTopology(), AbstractCellBasedSimulation< DIM >::UpdateCellPopulation(), and AbstractCellBasedSimulation< DIM >::WriteVisualizerSetupFile().
| bool AbstractCellBasedSimulation< DIM >::StoppingEventHasOccurred | ( | ) | [inline, protected, virtual] |
A child class can overload this if they want the simulation to stop based on certain conditions before the specified end time (for example, run until a crypt becomes monoclonal).
Definition at line 396 of file AbstractCellBasedSimulation.cpp.
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
| virtual void AbstractCellBasedSimulation< DIM >::UpdateCellLocationsAndTopology | ( | ) | [protected, pure virtual] |
Updates the locations and topology of cells, must be defined in subclasses.
For example this calculates forces and updates node positions in off lattice simulations.
Implemented in OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
| void AbstractCellBasedSimulation< DIM >::UpdateCellPopulation | ( | ) | [inline, protected, virtual] |
Calls the deaths, births and (if mUpdateCellPopulation is true) CellPopulation::Update() methods.
Reimplemented in OnLatticeSimulation< DIM >.
Definition at line 402 of file AbstractCellBasedSimulation.cpp.
References GenericEventHandler< 10, CellBasedEventHandler >::BeginEvent(), AbstractCellBasedSimulation< DIM >::DoCellBirth(), AbstractCellBasedSimulation< DIM >::DoCellRemoval(), GenericEventHandler< 10, CellBasedEventHandler >::EndEvent(), EXCEPTION, AbstractCellBasedSimulation< DIM >::mNumBirths, AbstractCellBasedSimulation< DIM >::mNumDeaths, AbstractCellBasedSimulation< DIM >::mrCellPopulation, and AbstractCellBasedSimulation< DIM >::mUpdateCellPopulation.
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
| virtual void AbstractCellBasedSimulation< DIM >::WriteVisualizerSetupFile | ( | ) | [inline, protected, virtual] |
Writes out special information about the mesh to the visualizer.
Reimplemented in OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Definition at line 155 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::Solve().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented in DeltaNotchOffLatticeSimulation< DIM >, OffLatticeSimulation< DIM >, OnLatticeSimulation< DIM >, VolumeTrackedOffLatticeSimulation< DIM >, CryptSimulation1d, CryptSimulation2d, OffLatticeSimulation< 1 >, and OffLatticeSimulation< 2 >.
Definition at line 68 of file AbstractCellBasedSimulation.hpp.
Member Data Documentation
std::vector<boost::shared_ptr<AbstractCellKiller<DIM> > > AbstractCellBasedSimulation< DIM >::mCellKillers [protected] |
List of cell killers.
Definition at line 139 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::AddCellKiller(), AbstractCellBasedSimulation< DIM >::DoCellRemoval(), AbstractCellBasedSimulation< DIM >::OutputSimulationSetup(), and AbstractCellBasedSimulation< DIM >::serialize().
bool AbstractCellBasedSimulation< DIM >::mDeleteCellPopulationInDestructor [protected] |
Whether to delete the cell population in the destructor.
Definition at line 112 of file AbstractCellBasedSimulation.hpp.
Referenced by CryptSimulation1d::CryptSimulation1d(), CryptSimulation2d::CryptSimulation2d(), and AbstractCellBasedSimulation< DIM >::~AbstractCellBasedSimulation().
double AbstractCellBasedSimulation< DIM >::mDt [protected] |
Time step.
Definition at line 103 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::GetDt(), OffLatticeSimulation< DIM >::OffLatticeSimulation(), OnLatticeSimulation< DIM >::OnLatticeSimulation(), AbstractCellBasedSimulation< DIM >::OutputSimulationParameters(), AbstractCellBasedSimulation< DIM >::serialize(), AbstractCellBasedSimulation< DIM >::SetDt(), AbstractCellBasedSimulation< DIM >::Solve(), OnLatticeSimulation< DIM >::UpdateCellLocationsAndTopology(), and OffLatticeSimulation< DIM >::UpdateNodePositions().
double AbstractCellBasedSimulation< DIM >::mEndTime [protected] |
Time to run the Solve() method up to.
Definition at line 106 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::OutputSimulationParameters(), AbstractCellBasedSimulation< DIM >::serialize(), AbstractCellBasedSimulation< DIM >::SetEndTime(), and AbstractCellBasedSimulation< DIM >::Solve().
bool AbstractCellBasedSimulation< DIM >::mInitialiseCells [protected] |
Whether to initialise the cells.
Definition at line 115 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::AbstractCellBasedSimulation(), and OnLatticeSimulation< DIM >::UpdateCellPopulation().
bool AbstractCellBasedSimulation< DIM >::mNoBirth [protected] |
Whether to run the simulation with no birth (defaults to false).
Definition at line 118 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::DoCellBirth(), AbstractCellBasedSimulation< DIM >::serialize(), and AbstractCellBasedSimulation< DIM >::SetNoBirth().
unsigned AbstractCellBasedSimulation< DIM >::mNumBirths [protected] |
Counts the number of births during the simulation.
Definition at line 133 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::GetNumBirths(), AbstractCellBasedSimulation< DIM >::serialize(), and AbstractCellBasedSimulation< DIM >::UpdateCellPopulation().
unsigned AbstractCellBasedSimulation< DIM >::mNumDeaths [protected] |
Counts the number of deaths during the simulation.
Definition at line 136 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::GetNumDeaths(), AbstractCellBasedSimulation< DIM >::serialize(), and AbstractCellBasedSimulation< DIM >::UpdateCellPopulation().
std::string AbstractCellBasedSimulation< DIM >::mOutputDirectory [protected] |
Output directory (a subfolder of tmp/[USERNAME]/testoutput).
Definition at line 124 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::GetOutputDirectory(), AbstractCellBasedSimulation< DIM >::serialize(), AbstractCellBasedSimulation< DIM >::SetOutputDirectory(), and AbstractCellBasedSimulation< DIM >::Solve().
CellBasedPdeHandler<DIM>* AbstractCellBasedSimulation< DIM >::mpCellBasedPdeHandler [protected] |
Pointer to a CellBasedPdeHandler object.
Definition at line 150 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::AfterSolve(), AbstractCellBasedSimulation< DIM >::GetCellBasedPdeHandler(), AbstractCellBasedSimulation< DIM >::OutputSimulationParameters(), AbstractCellBasedSimulation< DIM >::PostSolve(), AbstractCellBasedSimulation< DIM >::serialize(), AbstractCellBasedSimulation< DIM >::SetCellBasedPdeHandler(), AbstractCellBasedSimulation< DIM >::SetupSolve(), and AbstractCellBasedSimulation< DIM >::~AbstractCellBasedSimulation().
out_stream AbstractCellBasedSimulation< DIM >::mpVizSetupFile [protected] |
Visualiser setup file.
Definition at line 130 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::SetupSolve(), CryptSimulation2d::SetupWriteBetaCatenin(), AbstractCellBasedSimulation< DIM >::Solve(), OnLatticeSimulation< DIM >::WriteVisualizerSetupFile(), and OffLatticeSimulation< DIM >::WriteVisualizerSetupFile().
AbstractCellPopulation<DIM>& AbstractCellBasedSimulation< DIM >::mrCellPopulation [protected] |
Facade encapsulating cells in the cell population being simulated.
Definition at line 109 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::AbstractCellBasedSimulation(), OnLatticeSimulation< DIM >::AddCaUpdateRule(), OnLatticeSimulation< DIM >::AddPottsUpdateRule(), CryptSimulation2d::AfterSolve(), CryptSimulation2d::CalculateCellDivisionVector(), CryptSimulation1d::CalculateCellDivisionVector(), OffLatticeSimulation< DIM >::CalculateCellDivisionVector(), CryptSimulation1d::CryptSimulation1d(), CryptSimulation2d::CryptSimulation2d(), AbstractCellBasedSimulation< DIM >::DoCellBirth(), AbstractCellBasedSimulation< DIM >::DoCellRemoval(), AbstractCellBasedSimulation< DIM >::GetNodeLocation(), OnLatticeSimulation< DIM >::OutputAdditionalSimulationSetup(), CryptSimulation2d::OutputSimulationParameters(), AbstractCellBasedSimulation< DIM >::OutputSimulationSetup(), CryptSimulation2d::PostSolve(), VolumeTrackedOffLatticeSimulation< DIM >::PostSolve(), DeltaNotchOffLatticeSimulation< DIM >::PostSolve(), AbstractCellBasedSimulation< DIM >::rGetCellPopulation(), CryptSimulation2d::SetBottomCellAncestors(), CryptSimulation2d::SetupSolve(), AbstractCellBasedSimulation< DIM >::Solve(), OnLatticeSimulation< DIM >::UpdateCellLocationsAndTopology(), OffLatticeSimulation< DIM >::UpdateCellLocationsAndTopology(), OnLatticeSimulation< DIM >::UpdateCellPopulation(), AbstractCellBasedSimulation< DIM >::UpdateCellPopulation(), OffLatticeSimulation< DIM >::UpdateNodePositions(), CryptSimulation2d::WriteBetaCatenin(), OnLatticeSimulation< DIM >::WriteVisualizerSetupFile(), OffLatticeSimulation< DIM >::WriteVisualizerSetupFile(), and AbstractCellBasedSimulation< DIM >::~AbstractCellBasedSimulation().
unsigned AbstractCellBasedSimulation< DIM >::mSamplingTimestepMultiple [protected] |
The ratio of the number of actual timesteps to the number of timesteps at which results are written to file.
Definition at line 145 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::OutputSimulationParameters(), CryptSimulation2d::PostSolve(), AbstractCellBasedSimulation< DIM >::PostSolve(), AbstractCellBasedSimulation< DIM >::serialize(), AbstractCellBasedSimulation< DIM >::SetSamplingTimestepMultiple(), AbstractCellBasedSimulation< DIM >::Solve(), OnLatticeSimulation< DIM >::UpdateCellLocationsAndTopology(), and OffLatticeSimulation< DIM >::UpdateNodePositions().
std::string AbstractCellBasedSimulation< DIM >::mSimulationOutputDirectory [protected] |
Simulation Output directory either the same as mOutputDirectory or includes mOutputDirectory/results_from_time_[TIME].
Definition at line 127 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::OutputSimulationSetup(), AbstractCellBasedSimulation< DIM >::SetOutputDirectory(), OnLatticeSimulation< DIM >::SetupSolve(), OffLatticeSimulation< DIM >::SetupSolve(), AbstractCellBasedSimulation< DIM >::SetupSolve(), CryptSimulation2d::SetupWriteBetaCatenin(), and AbstractCellBasedSimulation< DIM >::Solve().
bool AbstractCellBasedSimulation< DIM >::mUpdateCellPopulation [protected] |
Whether to update the topology of the cell population at each time step (defaults to true).
Definition at line 121 of file AbstractCellBasedSimulation.hpp.
Referenced by AbstractCellBasedSimulation< DIM >::serialize(), AbstractCellBasedSimulation< DIM >::SetUpdateCellPopulationRule(), and AbstractCellBasedSimulation< DIM >::UpdateCellPopulation().
The documentation for this class was generated from the following files:
- cell_based/src/simulation/AbstractCellBasedSimulation.hpp
- cell_based/src/simulation/AbstractCellBasedSimulation.cpp
 1.6.3
1.6.3