#include <AbstractCardiacProblem.hpp>
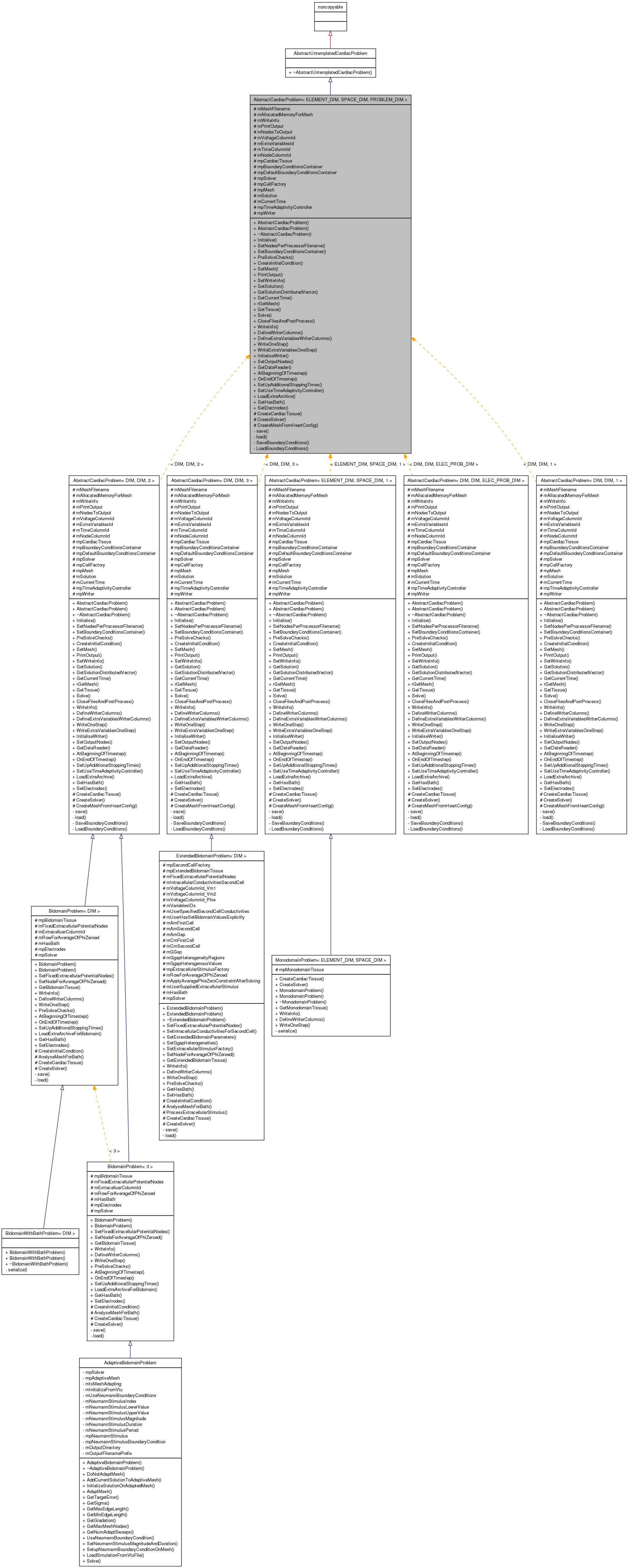
 Inheritance diagram for AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >:
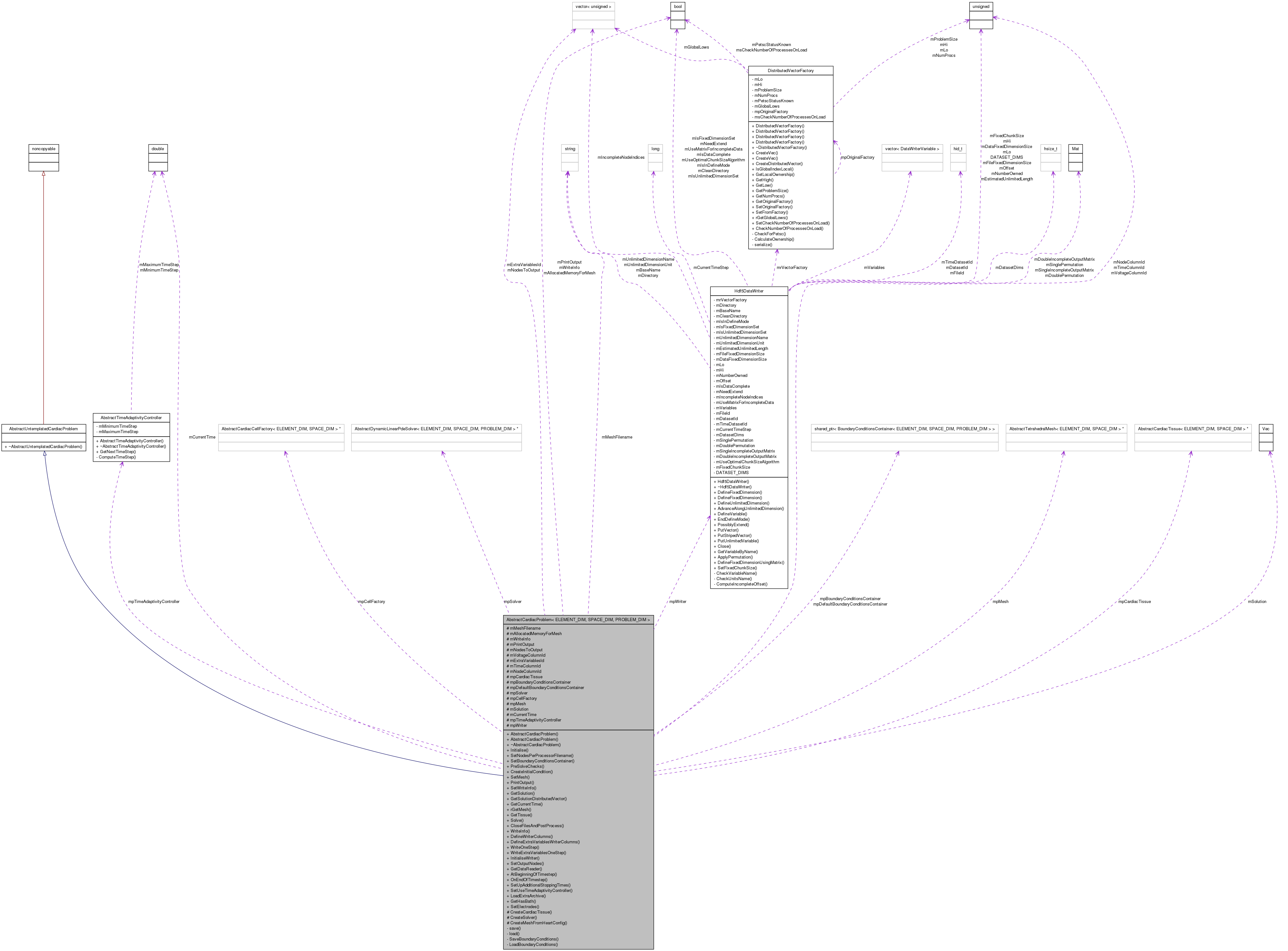
Inheritance diagram for AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >: Collaboration diagram for AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >:
Collaboration diagram for AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >:Detailed Description
template<unsigned ELEMENT_DIM, unsigned SPACE_DIM, unsigned PROBLEM_DIM>
class AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >
Base class for cardiac problems; contains code generic to both mono- and bidomain.
See tutorials for usage.
Definition at line 105 of file AbstractCardiacProblem.hpp.
Member Typedef Documentation
typedef boost::shared_ptr<BoundaryConditionsContainer<ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM> > AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::BccType [private] |
To save typing
Definition at line 112 of file AbstractCardiacProblem.hpp.
Constructor & Destructor Documentation
| AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::AbstractCardiacProblem | ( | AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM > * | pCellFactory | ) |
Constructor
- Parameters:
-
pCellFactory User defined cell factory which shows how the pde should create cells.
Definition at line 53 of file AbstractCardiacProblem.cpp.
References GenericEventHandler< 16, HeartEventHandler >::BeginEvent(), EXCEPTION, AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mNodesToOutput, and AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpCellFactory.
| AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::AbstractCardiacProblem | ( | ) |
Constructor used by archiving.
Definition at line 77 of file AbstractCardiacProblem.cpp.
| AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::~AbstractCardiacProblem | ( | ) | [virtual] |
Destructor
Definition at line 99 of file AbstractCardiacProblem.cpp.
References PetscTools::Destroy().
Member Function Documentation
| virtual void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::AtBeginningOfTimestep | ( | double | time | ) | [inline, virtual] |
Called at beginning of each time step in the main time-loop in Solve(). Empty implementation but can be overloaded by child classes.
- Parameters:
-
time the current time
Reimplemented in BidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 657 of file AbstractCardiacProblem.hpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::CloseFilesAndPostProcess | ( | ) |
Closes the files where the solution is stored and, if specified so (as it is by default), converts the output to Meshalyzer format by calling the WriteFilesUsingMesh method in the MeshalyzerWriter class.
- Note:
- This method is collective, and hence must be called by all processes.
Definition at line 557 of file AbstractCardiacProblem.cpp.
References GenericEventHandler< 16, HeartEventHandler >::BeginEvent(), GenericEventHandler< 16, HeartEventHandler >::EndEvent(), AbstractHdf5Converter< ELEMENT_DIM, SPACE_DIM >::GetSubdirectory(), HeartConfig::GetVisualizeWithMeshalyzer(), HeartConfig::Instance(), HeartConfig::Write(), and PostProcessingWriter< ELEMENT_DIM, SPACE_DIM >::WritePostProcessingFiles().
| virtual AbstractCardiacTissue<ELEMENT_DIM,SPACE_DIM>* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::CreateCardiacTissue | ( | ) | [protected, pure virtual] |
Subclasses must override this method to create a PDE object of the appropriate type.
This class will take responsibility for freeing the object when it is finished with.
Implemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, MonodomainProblem< ELEMENT_DIM, SPACE_DIM >, and BidomainProblem< 3 >.
| Vec AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::CreateInitialCondition | ( | ) | [virtual] |
This method sets the initial condition for the PDE by getting the voltages (V) from the cell models at the nodes.
If the problem dimension is two (Bidomain) the second variable (phi_e) is set to zero.
This is virtual so BidomainProblem can overwrite V to zero for bath nodes, if there are any.
- Todo:
- Perhaps this should be a method of AbstractCardiacTissue??
Reimplemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 255 of file AbstractCardiacProblem.cpp.
References DistributedVector::Begin(), DistributedVectorFactory::CreateDistributedVector(), DistributedVectorFactory::CreateVec(), DistributedVector::End(), and DistributedVector::Restore().
Referenced by BidomainProblem< DIM >::CreateInitialCondition().
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::CreateMeshFromHeartConfig | ( | ) | [protected, virtual] |
Subclasses must override this method to create a suitable mesh object.
Only needed if the subclass needs something other than a DistributedTetrahedralMesh.
This class will take responsibility for freeing the object when it is finished with.
Definition at line 207 of file AbstractCardiacProblem.cpp.
References HeartConfig::GetMeshPartitioning(), and HeartConfig::Instance().
| virtual AbstractDynamicLinearPdeSolver<ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM>* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::CreateSolver | ( | ) | [protected, pure virtual] |
Subclasses must override this method to create a suitable solver object.
This class will take responsibility for freeing the object when it is finished with.
Implemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, MonodomainProblem< ELEMENT_DIM, SPACE_DIM >, and BidomainProblem< 3 >.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::DefineExtraVariablesWriterColumns | ( | bool | extending | ) |
Define the user specified variables to be written to the primary results file
- Parameters:
-
extending whether we are extending an existing results file
Definition at line 653 of file AbstractCardiacProblem.cpp.
References HeartConfig::GetOutputVariables(), and HeartConfig::Instance().
Referenced by MonodomainProblem< ELEMENT_DIM, SPACE_DIM >::DefineWriterColumns(), ExtendedBidomainProblem< DIM >::DefineWriterColumns(), and BidomainProblem< DIM >::DefineWriterColumns().
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::DefineWriterColumns | ( | bool | extending | ) | [virtual] |
Define what variables are written to the primary results file.
- Parameters:
-
extending whether we are extending an existing results file
Reimplemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, MonodomainProblem< ELEMENT_DIM, SPACE_DIM >, and BidomainProblem< 3 >.
Definition at line 622 of file AbstractCardiacProblem.cpp.
References TimeStepper::EstimateTimeSteps(), and HeartConfig::Instance().
Referenced by MonodomainProblem< ELEMENT_DIM, SPACE_DIM >::DefineWriterColumns(), and BidomainProblem< DIM >::DefineWriterColumns().
| double AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::GetCurrentTime | ( | ) |
- Returns:
- the current time of the simulation
Definition at line 322 of file AbstractCardiacProblem.cpp.
| Hdf5DataReader AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::GetDataReader | ( | ) |
Create and return a data reader configured to read the results we've been outputting.
Definition at line 799 of file AbstractCardiacProblem.cpp.
References EXCEPTION, and HeartConfig::Instance().
| bool AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::GetHasBath | ( | ) | [virtual] |
Return whether there's bath defined in this problem
Reimplemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 809 of file AbstractCardiacProblem.cpp.
| Vec AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::GetSolution | ( | ) |
Get the final solution vector. This vector is distributed over all processes.
In case of Bidomain, this is of length 2*numNodes, and of the form (V_1, phi_1, V_2, phi_2, ......, V_N, phi_N). where V_j is the voltage at node j and phi_j is the extracellular potential at node j.
Use with caution since we don't want to alter the state of the PETSc vector.
Definition at line 310 of file AbstractCardiacProblem.cpp.
| DistributedVector AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::GetSolutionDistributedVector | ( | ) |
Get the solution vector, wrapped in a DistributedVector.
See also GetSolution.
Definition at line 316 of file AbstractCardiacProblem.cpp.
| AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM > * AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::GetTissue | ( | ) |
- Returns:
- the cardiac tissue object used
Definition at line 335 of file AbstractCardiacProblem.cpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::Initialise | ( | ) |
Initialise the system, once parameters have been set up.
Must be called before first calling Solve(). If loading from a checkpoint, do NOT call this method, as it can also be used to reset the problem to perform another simulation from time 0.
Definition at line 114 of file AbstractCardiacProblem.cpp.
References GenericEventHandler< 16, HeartEventHandler >::BeginEvent(), PetscTools::Destroy(), GenericEventHandler< 16, HeartEventHandler >::EndEvent(), EXCEPTION, HeartConfig::GetFibreLength(), HeartConfig::GetInterNodeSpace(), HeartConfig::GetMeshName(), HeartConfig::GetSheetDimensions(), Exception::GetShortMessage(), HeartConfig::GetSlabDimensions(), HeartConfig::Instance(), and NEVER_REACHED.
| bool AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::InitialiseWriter | ( | ) |
It creates and initialises the hdf writer from the Hdf5DataWriter class. It passes the output directory and file name to it. It is called by Solve(), if the output needs to be generated.
This method will try to open an existing .h5 file for extension if we are loading from an archive and one is present.
- Returns:
- whether the writer is outputting to an existing file.
Definition at line 726 of file AbstractCardiacProblem.cpp.
References RelativeTo::Absolute, PetscTools::Barrier(), EXCEPTION, FileFinder::Exists(), FileFinder::GetAbsolutePath(), OutputFileHandler::GetChasteTestOutputDirectory(), HeartConfig::GetOutputDirectory(), HeartConfig::GetOutputFilenamePrefix(), Hdf5DataReader::GetUnlimitedDimensionValues(), HeartConfig::Instance(), and HeartConfig::SetOutputUsingOriginalNodeOrdering().
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::load | ( | Archive & | archive, |
| const unsigned int | version | ||
| ) | [inline, private] |
Load the member variables.
- Parameters:
-
archive version
- Todo:
- #1317 code for saving/loading mSolution is PROBLEM_DIM specific, move it into the save/load methods fo Mono and BidomainProblem
- Todo:
- #1317 is there a reason we can't use PETSc's load/save vector functionality?
Reimplemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 212 of file AbstractCardiacProblem.hpp.
| BccType AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::LoadBoundaryConditions | ( | Archive & | archive, |
| AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > * | pMesh | ||
| ) | [inline, private] |
Serialization helper method to load a boundary conditions container.
- Parameters:
-
archive the archive to load from pMesh the mesh boundary conditions are to be defined on
- Returns:
- the loaded container
Definition at line 346 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load().
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::LoadExtraArchive | ( | Archive & | archive, |
| unsigned | version | ||
| ) |
Used when loading a set of archives written by a parallel simulation onto a single process. Loads data from the given process-specific archive (written by a non-master process) and merges it into our data.
- Parameters:
-
archive the archive to load version the archive file version
- Note:
- The process-specific archives currently contain the following data. If the layout changes, then this method will need to be altered, since it hard-codes knowledge of the order in which things are archived.
- (via mpMesh) DistributedVectorFactory*
- (via mpCardiacTissue LoadCardiacCells) DistributedVectorFactory*
- (via mpCardiacTissue LoadCardiacCells) number_of_cells and sequence of AbstractCardiacCell*, possibly with Purkinje interleaved
- (via mpCardiacTissue) DistributedVectorFactory*
- mpBoundaryConditionsContainer
- mpDefaultBoundaryConditionsContainer
- (if we're a BidomainProblem) stuff in BidomainProblem::LoadExtraArchiveForBidomain
- Todo:
- #1159 sanity check that the contents of p_bcc and mpBoundaryConditionsContainer match.
Definition at line 736 of file AbstractCardiacProblem.hpp.
References BidomainProblem< DIM >::LoadExtraArchiveForBidomain().
| virtual void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::OnEndOfTimestep | ( | double | time | ) | [inline, virtual] |
Called at end of each time step in the main time-loop in Solve(). Empty implementation but can be overloaded by child classes.
- Parameters:
-
time the current time
Reimplemented in BidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 667 of file AbstractCardiacProblem.hpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::PreSolveChecks | ( | ) | [virtual] |
Performs a series of checks before solving. It checks whether the cardiac pde has been defined, whether the simulation time is greater than zero and whether the output directory is specified (or the output is set not to be produced). It throws exceptions if any of the above checks fails.
- Todo:
- remove magic number? (#1884)
Reimplemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 219 of file AbstractCardiacProblem.cpp.
References EXCEPTION, HeartConfig::GetPdeTimeStep(), HeartConfig::GetSimulationDuration(), and HeartConfig::Instance().
Referenced by ExtendedBidomainProblem< DIM >::PreSolveChecks(), and BidomainProblem< DIM >::PreSolveChecks().
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::PrintOutput | ( | bool | rPrintOutput | ) |
Set whether the simulation will generate results files.
- Parameters:
-
rPrintOutput
Definition at line 298 of file AbstractCardiacProblem.cpp.
| AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > & AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::rGetMesh | ( | ) |
- Returns:
- the mesh used
Definition at line 328 of file AbstractCardiacProblem.cpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::save | ( | Archive & | archive, |
| const unsigned int | version | ||
| ) | const [inline, private] |
Save the member variables.
- Parameters:
-
archive version
- Todo:
- #1317 code for saving/loading mSolution is PROBLEM_DIM specific, move it into the save/load methods for Mono and BidomainProblem
- Todo:
- #1369
Reimplemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 125 of file AbstractCardiacProblem.hpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SaveBoundaryConditions | ( | Archive & | archive, |
| AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > * | pMesh, | ||
| BccType | pBcc | ||
| ) | const [inline, private] |
Serialization helper method to save a boundary conditions container.
- Parameters:
-
archive the archive to save to pMesh the mesh boundary conditions are defined on pBcc the container to save
Definition at line 331 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::save().
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetBoundaryConditionsContainer | ( | BccType | pBcc | ) |
Set the boundary conditions container.
- Parameters:
-
pBcc is a pointer to a boundary conditions container
Definition at line 213 of file AbstractCardiacProblem.cpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetElectrodes | ( | ) | [virtual] |
Set an electrode object (which provides boundary conditions). Only valid if there is a bath.
Reimplemented in BidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 814 of file AbstractCardiacProblem.cpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetMesh | ( | AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > * | pMesh | ) |
This only needs to be called if a mesh filename has not been set.
- Parameters:
-
pMesh the mesh object to use
Definition at line 285 of file AbstractCardiacProblem.cpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetNodesPerProcessorFilename | ( | const std::string & | rFilename | ) |
Set a file from which the nodes for each processor are read
- Parameters:
-
rFilename
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetOutputNodes | ( | std::vector< unsigned > & | rNodesToOutput | ) |
Specifies which nodes in the mesh to output.
- Parameters:
-
rNodesToOutput is a reference to a vector with the indexes of the nodes where the output is desired. If empty, the output will be for all the nodes in the mesh.
Definition at line 793 of file AbstractCardiacProblem.cpp.
| virtual void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetUpAdditionalStoppingTimes | ( | std::vector< double > & | rAdditionalStoppingTimes | ) | [inline, virtual] |
Allow subclasses to define additional 'stopping times' for the printing time step loop. This allows bidomain simulations to specify exactly when the Electrodes should be turned on or off.
- Parameters:
-
rAdditionalStoppingTimes to be filled in with the additional stopping times
Reimplemented in BidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 677 of file AbstractCardiacProblem.hpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetUseTimeAdaptivityController | ( | bool | useAdaptivity, |
| AbstractTimeAdaptivityController * | pController = NULL |
||
| ) |
- Todo:
- #1704 add default adaptivity controller and allow the user just to call with true
Set whether (or not) to use a time adaptivity controller
- Parameters:
-
useAdaptivity whether to use adaptivity pController The controller (only relevant if useAdaptivity==true)
Definition at line 341 of file AbstractCardiacProblem.cpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::SetWriteInfo | ( | bool | writeInfo = true | ) |
Set whether extra info will be written to stdout during computation.
- Parameters:
-
writeInfo
Definition at line 304 of file AbstractCardiacProblem.cpp.
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::Solve | ( | ) |
First performs some checks by calling the PreSolveChecks method. It creates an solver to which it passes the boundary conditions specified by the user (otherwise it passes the defauls bcc). It then calls the Solve method on the solver class. It also handles the output, if necessary.
- Note:
- This method is collective, and hence must be called by all processes.
Reimplemented in AdaptiveBidomainProblem.
Definition at line 358 of file AbstractCardiacProblem.cpp.
References TimeStepper::AdvanceOneTimeStep(), GenericEventHandler< 16, HeartEventHandler >::BeginEvent(), PetscTools::Destroy(), GenericEventHandler< 16, HeartEventHandler >::EndEvent(), TimeStepper::GetNextTime(), HeartConfig::GetOutputDirectory(), TimeStepper::GetTime(), HeartConfig::Instance(), TimeStepper::IsTimeAtEnd(), ProgressReporter::PrintFinalising(), PetscTools::ReplicateException(), GenericEventHandler< 16, HeartEventHandler >::Reset(), and ProgressReporter::Update().
| void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::WriteExtraVariablesOneStep | ( | ) |
Write one timestep of output data for the extra variables to the primary results file.
Definition at line 689 of file AbstractCardiacProblem.cpp.
References DistributedVector::Begin(), PetscTools::Destroy(), DistributedVector::End(), HeartConfig::GetOutputVariables(), HeartConfig::Instance(), and DistributedVector::Restore().
Referenced by MonodomainProblem< ELEMENT_DIM, SPACE_DIM >::WriteOneStep(), ExtendedBidomainProblem< DIM >::WriteOneStep(), and BidomainProblem< DIM >::WriteOneStep().
| virtual void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::WriteInfo | ( | double | time | ) | [pure virtual] |
Write informative details about the progress of the simulation to standard output.
Implemented only in subclasses.
- Parameters:
-
time the current time
Implemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, MonodomainProblem< ELEMENT_DIM, SPACE_DIM >, and BidomainProblem< 3 >.
| virtual void AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::WriteOneStep | ( | double | time, |
| Vec | voltageVec | ||
| ) | [pure virtual] |
Write one timestep of output data to the primary results file.
- Parameters:
-
time the current time voltageVec the solution vector to write
Implemented in BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, MonodomainProblem< ELEMENT_DIM, SPACE_DIM >, and BidomainProblem< 3 >.
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented in BidomainProblem< DIM >, BidomainWithBathProblem< DIM >, ExtendedBidomainProblem< DIM >, MonodomainProblem< ELEMENT_DIM, SPACE_DIM >, and BidomainProblem< 3 >.
Definition at line 116 of file AbstractCardiacProblem.hpp.
friend class CardiacElectroMechanicsProblem [friend] |
CardiacElectroMechanicsProblem needs access to mpWriter.
Definition at line 446 of file AbstractCardiacProblem.hpp.
Member Data Documentation
bool AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mAllocatedMemoryForMesh [protected] |
Whether this problem class has created the mesh itself, as opposed to being given it
Definition at line 369 of file AbstractCardiacProblem.hpp.
double AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mCurrentTime [protected] |
The current simulation time.
This is used to be able to restart simulations at a point other than time zero, either because of repeated calls to Solve (with increased simulation duration) or because of restarting from a checkpoint.
Definition at line 412 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
std::vector<unsigned> AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mExtraVariablesId [protected] |
List of extra variables to be written to HDF5 file
Definition at line 381 of file AbstractCardiacProblem.hpp.
std::string AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mMeshFilename [protected] |
Meshes can be read from file or instantiated and passed directly to this class, this is for the former
Definition at line 366 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
unsigned AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mNodeColumnId [protected] |
Used by the writer
Definition at line 385 of file AbstractCardiacProblem.hpp.
std::vector<unsigned> AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mNodesToOutput [protected] |
If only outputing voltage for selected nodes, which nodes to output at
Definition at line 376 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::AbstractCardiacProblem(), AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
BccType AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpBoundaryConditionsContainer [protected] |
Boundary conditions container used in the simulation
Definition at line 391 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
AbstractCardiacTissue<ELEMENT_DIM,SPACE_DIM>* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpCardiacTissue [protected] |
The monodomain or bidomain pde
Definition at line 388 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
AbstractCardiacCellFactory<ELEMENT_DIM,SPACE_DIM>* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpCellFactory [protected] |
The cell factory creates the cells for each node
Definition at line 397 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::AbstractCardiacProblem().
BccType AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpDefaultBoundaryConditionsContainer [protected] |
It is convenient to also have a separate variable for default (zero-Neumann) boundary conditions
Definition at line 393 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
AbstractTetrahedralMesh<ELEMENT_DIM,SPACE_DIM>* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpMesh [protected] |
The mesh. Can either by passed in, or the mesh filename can be set
Definition at line 399 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
bool AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mPrintOutput [protected] |
Whether to write any output at all
Definition at line 373 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
AbstractDynamicLinearPdeSolver<ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM>* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpSolver [protected] |
The PDE solver
Reimplemented in AdaptiveBidomainProblem, BidomainProblem< DIM >, ExtendedBidomainProblem< DIM >, and BidomainProblem< 3 >.
Definition at line 395 of file AbstractCardiacProblem.hpp.
AbstractTimeAdaptivityController* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpTimeAdaptivityController [protected] |
Adaptivity controller (defaults to NULL).
Definition at line 415 of file AbstractCardiacProblem.hpp.
Hdf5DataWriter* AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mpWriter [protected] |
The object to use to write results to disk.
Definition at line 450 of file AbstractCardiacProblem.hpp.
Vec AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mSolution [protected] |
The current solution vector, of the form [V_0 .. V_N ] for monodomain and [V_0 phi_0 .. V_N phi_N] for bidomain
Definition at line 403 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
unsigned AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mTimeColumnId [protected] |
Used by the writer
Definition at line 383 of file AbstractCardiacProblem.hpp.
unsigned AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mVoltageColumnId [protected] |
Used by the writer
Definition at line 379 of file AbstractCardiacProblem.hpp.
bool AbstractCardiacProblem< ELEMENT_DIM, SPACE_DIM, PROBLEM_DIM >::mWriteInfo [protected] |
Whether to print some statistics (max/min voltage) to screen during the simulation
Definition at line 371 of file AbstractCardiacProblem.hpp.
Referenced by AbstractCardiacProblem< DIM, DIM, 1 >::load(), and AbstractCardiacProblem< DIM, DIM, 1 >::save().
The documentation for this class was generated from the following files:
- heart/src/problem/AbstractCardiacProblem.hpp
- heart/src/problem/AbstractCardiacProblem.cpp