#include <AbstractCardiacTissue.hpp>
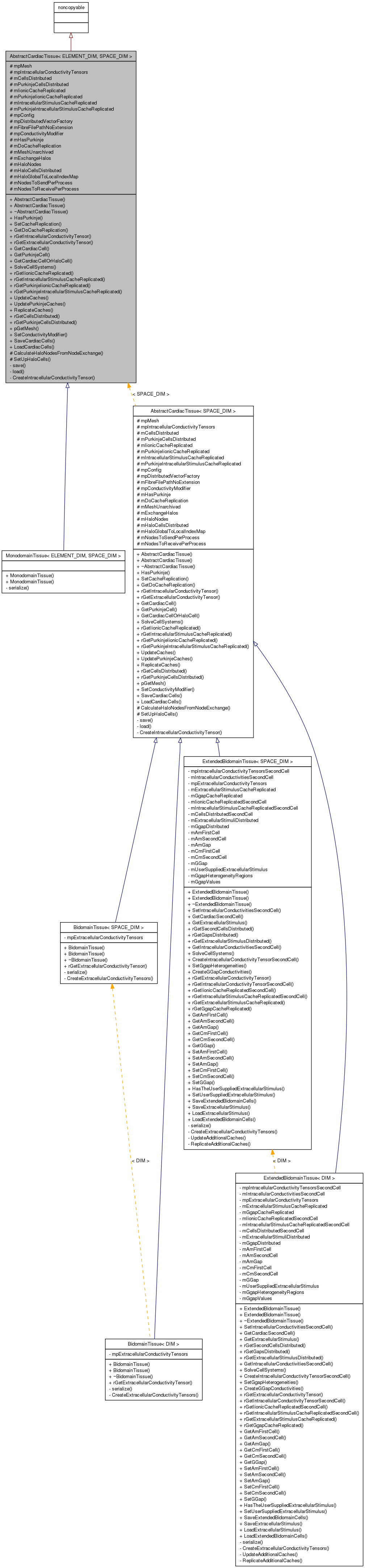
 Inheritance diagram for AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >:
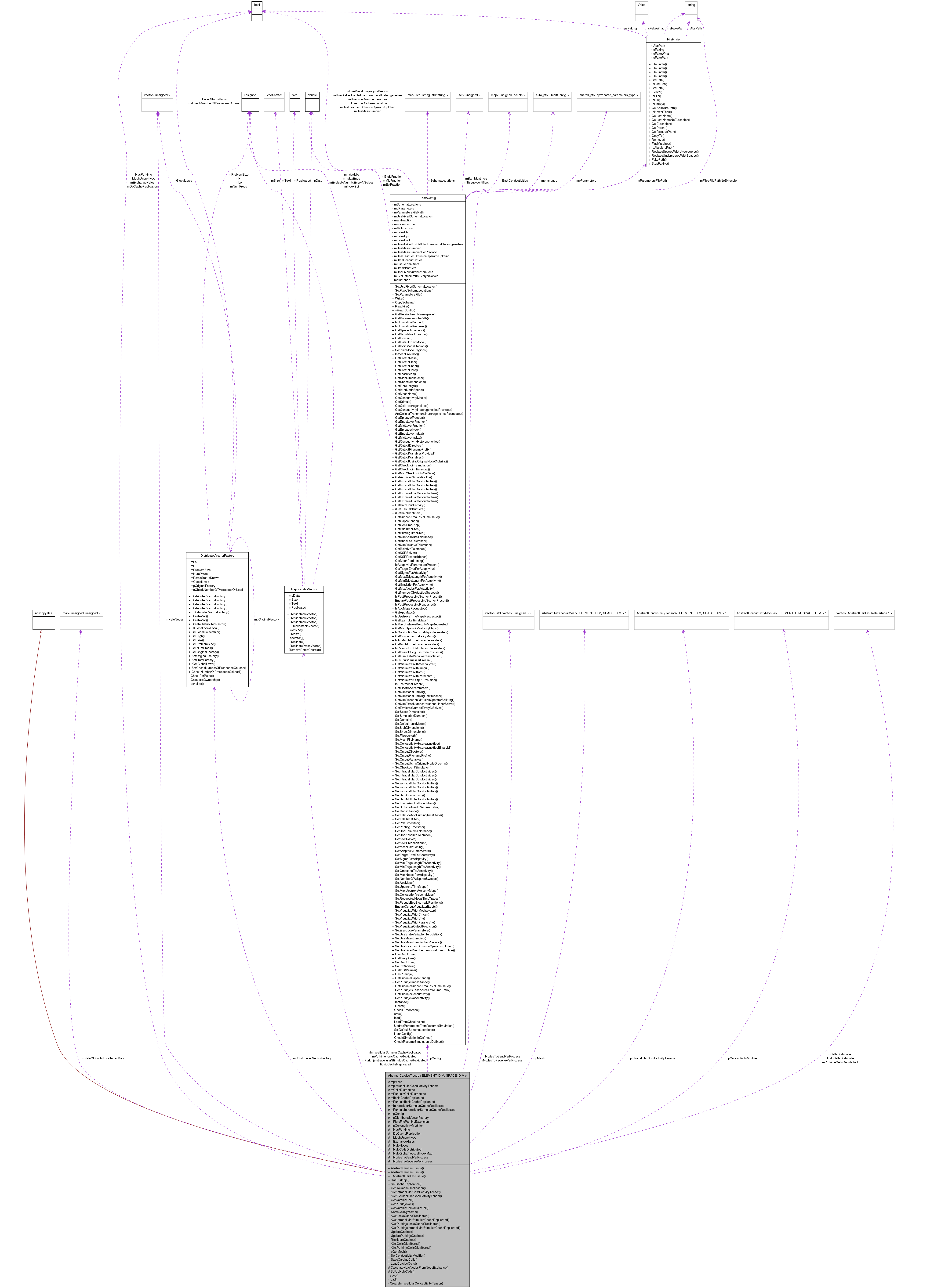
Inheritance diagram for AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >: Collaboration diagram for AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >:
Collaboration diagram for AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >:Detailed Description
template<unsigned ELEMENT_DIM, unsigned SPACE_DIM = ELEMENT_DIM>
class AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >
Class containing "tissue-like" functionality used in monodomain and bidomain problems.
Contains the cardiac cells (ODE systems for each node of the mesh) and conductivity tensors (dependent on fibre directions).
Also contains knowledge of parallelisation in the form of the distributed vector factory. This class deals with created a distributed vector of cells, and getting the ionic current and stimuli from these cells and putting them in replicated arrays for the PDE solvers to call.
Definition at line 78 of file AbstractCardiacTissue.hpp.
Constructor & Destructor Documentation
| AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue | ( | AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM > * | pCellFactory, |
| bool | exchangeHalos = false |
||
| ) |
This constructor is called from the Initialise() method of the CardiacProblem class. It creates all the cell objects, and sets up the conductivities.
Note that pCellFactory contains a pointer to the mesh
- Parameters:
-
pCellFactory factory to use to create cardiac cells. If this is actually an AbstractPurkinjeCellFactory it creates purkinje cells. exchangeHalos used in state-variable interpolation. Defaults to false.
Definition at line 49 of file AbstractCardiacTissue.cpp.
References GenericEventHandler< 16, HeartEventHandler >::BeginEvent(), AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM >::CreateCardiacCellForNode(), AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::CreateIntracellularConductivityTensor(), AbstractPurkinjeCellFactory< ELEMENT_DIM, SPACE_DIM >::CreatePurkinjeCellForNode(), GenericEventHandler< 16, HeartEventHandler >::EndEvent(), AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM >::FinaliseCellCreation(), AbstractPurkinjeCellFactory< ELEMENT_DIM, SPACE_DIM >::FinalisePurkinjeCellCreation(), DistributedVectorFactory::GetHigh(), DistributedVectorFactory::GetLocalOwnership(), DistributedVectorFactory::GetLow(), AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM >::GetMesh(), HeartConfig::GetMeshName(), AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM >::GetNumberOfCells(), HeartConfig::Instance(), PetscTools::IsSequential(), AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mCellsDistributed, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mExchangeHalos, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mFibreFilePathNoExtension, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mHasPurkinje, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mIionicCacheReplicated, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mIntracellularStimulusCacheReplicated, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mpDistributedVectorFactory, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mPurkinjeCellsDistributed, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mPurkinjeIionicCacheReplicated, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mPurkinjeIntracellularStimulusCacheReplicated, PetscTools::ReplicateException(), ReplicatableVector::Resize(), and AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::SetUpHaloCells().
| AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue | ( | AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > * | pMesh | ) |
This constructor is called by the archiver only.
- Parameters:
-
pMesh a pointer to the AbstractTetrahedral mesh.
Definition at line 162 of file AbstractCardiacTissue.cpp.
References AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::CreateIntracellularConductivityTensor(), ArchiveLocationInfo::GetArchiveDirectory(), ArchiveLocationInfo::GetMeshFilename(), DistributedVectorFactory::GetProblemSize(), AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mFibreFilePathNoExtension, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mIionicCacheReplicated, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mIntracellularStimulusCacheReplicated, AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mpDistributedVectorFactory, and ReplicatableVector::Resize().
| AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::~AbstractCardiacTissue | ( | ) | [virtual] |
Virtual destructor
Definition at line 178 of file AbstractCardiacTissue.cpp.
Member Function Documentation
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::CalculateHaloNodesFromNodeExchange | ( | ) | [protected] |
If the mesh is a tetrahedral mesh then all elements and nodes are known. The halo nodes to the ones which are actually used as cardiac cells must be calculated explicitly.
Definition at line 403 of file AbstractCardiacTissue.cpp.
References PetscTools::GetNumProcs().
Referenced by AbstractCardiacTissue< SPACE_DIM >::load().
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::CreateIntracellularConductivityTensor | ( | ) | [private] |
Convenience method for intracellular conductivity tensor creation
- Todo:
- #1316 Create a class defining constant tensors to be used when no fibre orientation is provided.
- Todo:
- #1316 Create a class defining constant tensors to be used when no fibre orientation is provided.
Definition at line 221 of file AbstractCardiacTissue.cpp.
References RelativeTo::AbsoluteOrCwd, GenericEventHandler< 16, HeartEventHandler >::BeginEvent(), GenericEventHandler< 16, HeartEventHandler >::EndEvent(), FileFinder::Exists(), HeartConfig::GetConductivityHeterogeneities(), HeartConfig::Instance(), NEVER_REACHED, and PetscTools::ReplicateException().
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue().
| AbstractCardiacCellInterface * AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::GetCardiacCell | ( | unsigned | globalIndex | ) |
Get a pointer to a cell, indexed by the global node index.
- Note:
- Should only called by the process owning the cell - triggers an assertion otherwise.
- Parameters:
-
globalIndex global node index for which to retrieve a cell
Definition at line 365 of file AbstractCardiacTissue.cpp.
Referenced by AdaptiveBidomainProblem::AddCurrentSolutionToAdaptiveMesh(), CardiacSimulation::CreateAndRun(), and AdaptiveBidomainProblem::InitializeSolutionOnAdaptedMesh().
| AbstractCardiacCellInterface * AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::GetCardiacCellOrHaloCell | ( | unsigned | globalIndex | ) |
Get a pointer to a halo cell, indexed by the global node index.
- Note:
- Should only called by the process halo owning the cell - triggers an assertion otherwise.
- Parameters:
-
globalIndex global node index for which to retrieve a cell
Definition at line 382 of file AbstractCardiacTissue.cpp.
References EXCEPTION, and PetscTools::GetMyRank().
| bool AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::GetDoCacheReplication | ( | ) |
Get whether or not to replicate the caches across all processors.
- Returns:
- mDoCacheReplication - true if the cache needs to be replicated
Definition at line 339 of file AbstractCardiacTissue.cpp.
| AbstractCardiacCellInterface * AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::GetPurkinjeCell | ( | unsigned | globalIndex | ) |
Get a pointer to a Purkinje cell, indexed by the global node index.
- Note:
- Should only called by the process owning the cell - triggers an assertion otherwise.
- Parameters:
-
globalIndex global node index for which to retrieve a cell
Definition at line 373 of file AbstractCardiacTissue.cpp.
References EXCEPT_IF_NOT.
| bool AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::HasPurkinje | ( | ) |
Determine whether this tissue contains Purkinje fibres.
Definition at line 215 of file AbstractCardiacTissue.cpp.
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::load | ( | Archive & | archive, |
| const unsigned int | version | ||
| ) | [inline, private] |
Unarchive the member variables.
- Parameters:
-
archive version
Definition at line 169 of file AbstractCardiacTissue.hpp.
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::LoadCardiacCells | ( | Archive & | archive, |
| const unsigned int | version | ||
| ) | [inline] |
Load our tissue from an archive.
Handles the checkpoint migration case, deleting loaded cells immediately if they are not local to this process.
Also loads halo cells if we're doing halo exchange, by using the non-local cells from the archive.
- Parameters:
-
archive the process-specific archive to load from version archive version
- Todo:
- #1199 test this
Definition at line 567 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load().
| const AbstractTetrahedralMesh< ELEMENT_DIM, SPACE_DIM > * AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::pGetMesh | ( | ) | const |
Returns a pointer to the mesh object
- Returns:
- pointer to mesh object
Definition at line 731 of file AbstractCardiacTissue.cpp.
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::ReplicateCaches | ( | ) |
Replicate the Iionic and intracellular stimulus caches.
Definition at line 701 of file AbstractCardiacTissue.cpp.
| const std::vector< AbstractCardiacCellInterface * > & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetCellsDistributed | ( | ) | const |
Returns a reference to the vector of distributed cells. Needed for archiving.
Definition at line 718 of file AbstractCardiacTissue.cpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::SaveCardiacCells().
| const c_matrix< double, SPACE_DIM, SPACE_DIM > & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetExtracellularConductivityTensor | ( | unsigned | elementIndex | ) | [virtual] |
Get the extracellular conductivity tensor for the given element (this throws an exception in this abstract class since monodomain don't have extracellular ones) it is overridden in the BidomainTissue.
- Parameters:
-
elementIndex index of the element of interest
Reimplemented in BidomainTissue< SPACE_DIM >, ExtendedBidomainTissue< SPACE_DIM >, BidomainTissue< DIM >, and ExtendedBidomainTissue< DIM >.
Definition at line 359 of file AbstractCardiacTissue.cpp.
References EXCEPTION.
| ReplicatableVector & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetIionicCacheReplicated | ( | ) |
Get the entire ionic current cache
Definition at line 660 of file AbstractCardiacTissue.cpp.
| const c_matrix< double, SPACE_DIM, SPACE_DIM > & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetIntracellularConductivityTensor | ( | unsigned | elementIndex | ) |
Get the intracellular conductivity tensor for the given element
- Parameters:
-
elementIndex index of the element of interest
Definition at line 345 of file AbstractCardiacTissue.cpp.
| ReplicatableVector & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetIntracellularStimulusCacheReplicated | ( | ) |
Get the entire stimulus current cache
Definition at line 666 of file AbstractCardiacTissue.cpp.
| const std::vector< AbstractCardiacCellInterface * > & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetPurkinjeCellsDistributed | ( | ) | const |
Returns a reference to the vector of distributed Purkinje cells. Needed for archiving.
Definition at line 724 of file AbstractCardiacTissue.cpp.
References EXCEPT_IF_NOT.
Referenced by AbstractCardiacTissue< SPACE_DIM >::SaveCardiacCells().
| ReplicatableVector & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetPurkinjeIionicCacheReplicated | ( | ) |
Get the entire Purkinje ionic current cache
Definition at line 672 of file AbstractCardiacTissue.cpp.
References EXCEPT_IF_NOT.
| ReplicatableVector & AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::rGetPurkinjeIntracellularStimulusCacheReplicated | ( | ) |
Get the entire Purkinje stimulus current cache
Definition at line 679 of file AbstractCardiacTissue.cpp.
References EXCEPT_IF_NOT.
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::save | ( | Archive & | archive, |
| const unsigned int | version | ||
| ) | const [inline, private] |
Archive the member variables.
- Parameters:
-
archive version
Definition at line 93 of file AbstractCardiacTissue.hpp.
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::SaveCardiacCells | ( | Archive & | archive, |
| const unsigned int | version | ||
| ) | const [inline] |
Save our tissue to an archive.
Writes:
- mpDistributedVectorFactory
- number of cells on this process
- each cell pointer in turn, interleaved with Purkinje cells if present
- Parameters:
-
archive the process-specific archive to write cells to. version
Definition at line 526 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::save().
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::SetCacheReplication | ( | bool | doCacheReplication | ) |
Set whether or not to replicate the caches across all processors.
See also mDoCacheReplication.
- Parameters:
-
doCacheReplication - true if the cache needs to be replicated
Definition at line 333 of file AbstractCardiacTissue.cpp.
Referenced by MonodomainSolver< ELEMENT_DIM, SPACE_DIM >::MonodomainSolver(), and OperatorSplittingMonodomainSolver< ELEMENT_DIM, SPACE_DIM >::OperatorSplittingMonodomainSolver().
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::SetConductivityModifier | ( | AbstractConductivityModifier< ELEMENT_DIM, SPACE_DIM > * | pModifier | ) |
Set a modifier class which will be used to modifier a conductivity obtained from mpIntracellularConductivityTensors when rGetIntracellularConductivityTensor() is called. For example, it is required when conductivities become deformation-dependent.
- Parameters:
-
pModifier Pointer to the concrete modifier class
Definition at line 737 of file AbstractCardiacTissue.cpp.
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::SetUpHaloCells | ( | AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM > * | pCellFactory | ) | [protected] |
If mExchangeHalos is true, this method calls CalculateHaloNodesFromNodeExchange and sets up the halo cell data structures mHaloCellsDistributed and mHaloGlobalToLocalIndexMap.
- Parameters:
-
pCellFactory cell factory to use to create halo cells
Definition at line 416 of file AbstractCardiacTissue.cpp.
References AbstractCardiacCellFactory< ELEMENT_DIM, SPACE_DIM >::CreateCardiacCellForNode(), PetscTools::ReplicateException(), and AbstractCardiacCellInterface::SetUsedInTissueSimulation().
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue().
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::SolveCellSystems | ( | Vec | existingSolution, |
| double | time, | ||
| double | nextTime, | ||
| bool | updateVoltage = false |
||
| ) | [virtual] |
Integrate the cell ODEs and update ionic current etc for each of the cells, between the two times provided.
- Parameters:
-
existingSolution the current voltage solution vector time the current simulation time nextTime when to simulate the cells until updateVoltage whether to also solve for the voltage (generally false, true for operator splitting methods). Defaults to false
- Todo:
- #2017 This code will be needed in the future, not being covered now. Add a test which covers this, e.g. by doing a simulation with a bad stimulus for one of the Purkinje cells - stimulating with the wrong choice of sign say.
Reimplemented in ExtendedBidomainTissue< SPACE_DIM >, and ExtendedBidomainTissue< DIM >.
Definition at line 465 of file AbstractCardiacTissue.cpp.
References DistributedVector::Begin(), GenericEventHandler< 16, HeartEventHandler >::BeginEvent(), DistributedVector::End(), GenericEventHandler< 16, HeartEventHandler >::EndEvent(), PetscTools::GetMyRank(), AbstractCardiacCellInterface::GetNumberOfStateVariables(), PetscTools::GetNumProcs(), PetscVecTools::GetSize(), AbstractCardiacCellInterface::GetStdVecStateVariables(), NEVER_REACHED, PetscTools::ReplicateException(), DistributedVector::Restore(), and AbstractCardiacCellInterface::SetStateVariables().
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::UpdateCaches | ( | unsigned | globalIndex, |
| unsigned | localIndex, | ||
| double | nextTime | ||
| ) |
Update the Iionic and intracellular stimulus caches.
- Parameters:
-
globalIndex global index of the entry to update localIndex local index of the entry to update nextTime the next PDE time point, at which to evaluate the stimulus current
Definition at line 686 of file AbstractCardiacTissue.cpp.
| void AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::UpdatePurkinjeCaches | ( | unsigned | globalIndex, |
| unsigned | localIndex, | ||
| double | nextTime | ||
| ) |
Update the Iionic and intracellular stimulus caches for Purkinje cells.
- Parameters:
-
globalIndex global index of the entry to update localIndex local index of the entry to update nextTime the next PDE time point, at which to evaluate the stimulus current
Definition at line 693 of file AbstractCardiacTissue.cpp.
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented in BidomainTissue< SPACE_DIM >, ExtendedBidomainTissue< SPACE_DIM >, MonodomainTissue< ELEMENT_DIM, SPACE_DIM >, BidomainTissue< DIM >, and ExtendedBidomainTissue< DIM >.
Definition at line 83 of file AbstractCardiacTissue.hpp.
Member Data Documentation
std::vector< AbstractCardiacCellInterface* > AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mCellsDistributed [protected] |
The vector of cells. Distributed.
Definition at line 242 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue(), and AbstractCardiacTissue< SPACE_DIM >::LoadCardiacCells().
bool AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mDoCacheReplication [protected] |
Whether we need to replicate the caches.
When doing matrix-based RHS assembly, we only actually need information from cells/nodes local to the processor, so replicating the caches is an unnecessary communication overhead.
Defaults to true.
Definition at line 308 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load(), and AbstractCardiacTissue< SPACE_DIM >::save().
bool AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mExchangeHalos [protected] |
Whether to exchange cell models across the halo boundaries. Used in state variable interpolation.
Definition at line 319 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue(), AbstractCardiacTissue< SPACE_DIM >::load(), and AbstractCardiacTissue< SPACE_DIM >::save().
std::string AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mFibreFilePathNoExtension [protected] |
Path to the location of the fibre file without extension.
Definition at line 287 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue(), and AbstractCardiacTissue< SPACE_DIM >::save().
std::vector< AbstractCardiacCellInterface* > AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mHaloCellsDistributed [protected] |
The vector of halo cells. Distributed.
Definition at line 325 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load(), and AbstractCardiacTissue< SPACE_DIM >::LoadCardiacCells().
std::map<unsigned, unsigned> AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mHaloGlobalToLocalIndexMap [protected] |
Map of global to local indices for halo nodes.
Definition at line 328 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load(), and AbstractCardiacTissue< SPACE_DIM >::LoadCardiacCells().
std::vector<unsigned> AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mHaloNodes [protected] |
Vector of halo node indices for current process
Definition at line 322 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load().
bool AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mHasPurkinje [protected] |
Whether this tissue has any Purkinje cells.
Definition at line 297 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue(), AbstractCardiacTissue< SPACE_DIM >::load(), AbstractCardiacTissue< SPACE_DIM >::LoadCardiacCells(), and AbstractCardiacTissue< SPACE_DIM >::save().
ReplicatableVector AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mIionicCacheReplicated [protected] |
Cache containing all the ionic currents for each node, replicated over all processes.
Definition at line 251 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue().
ReplicatableVector AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mIntracellularStimulusCacheReplicated [protected] |
Cache containing all the stimulus currents for each node, replicated over all processes.
Definition at line 263 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue().
bool AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mMeshUnarchived [protected] |
Whether the mesh was unarchived or got from elsewhere.
Definition at line 313 of file AbstractCardiacTissue.hpp.
std::vector<std::vector<unsigned> > AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mNodesToReceivePerProcess [protected] |
A vector which will be of size GetNumProcs() for information to receive for process i.
Definition at line 341 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load().
std::vector<std::vector<unsigned> > AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mNodesToSendPerProcess [protected] |
A vector which will be of size GetNumProcs() where each internal vector except i=GetMyRank() contains an ordered list of indices of nodes to send to process i during data exchange
Definition at line 335 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load().
AbstractConductivityModifier<ELEMENT_DIM,SPACE_DIM>* AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mpConductivityModifier [protected] |
This class, if not NULL, will be used to modify the conductivity that is obtained from mpIntracellularConductivityTensors when rGetIntracellularConductivityTensor() is called. For example, it is required when conductivities become deformation dependent.
Definition at line 294 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load().
HeartConfig* AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mpConfig [protected] |
Local pointer to the HeartConfig singleton instance, for convenience.
Definition at line 272 of file AbstractCardiacTissue.hpp.
DistributedVectorFactory* AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mpDistributedVectorFactory [protected] |
Local pointer to the distributed vector factory associated with the mesh object used.
Used to retrieve node ownership range when needed.
NB: This is set from mpMesh->GetDistributedVectorFactory() and thus always equal to that. We never assume ownership of the object.
Definition at line 282 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue(), AbstractCardiacTissue< SPACE_DIM >::load(), AbstractCardiacTissue< SPACE_DIM >::save(), and AbstractCardiacTissue< SPACE_DIM >::SaveCardiacCells().
AbstractConductivityTensors<ELEMENT_DIM,SPACE_DIM>* AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mpIntracellularConductivityTensors [protected] |
Intracellular conductivity tensors. Not archived, since it's loaded from the HeartConfig singleton.
Definition at line 239 of file AbstractCardiacTissue.hpp.
AbstractTetrahedralMesh<ELEMENT_DIM,SPACE_DIM>* AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mpMesh [protected] |
It's handy to keep a pointer to the mesh object
Definition at line 235 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< SPACE_DIM >::load(), AbstractCardiacTissue< SPACE_DIM >::LoadCardiacCells(), and AbstractCardiacTissue< SPACE_DIM >::save().
std::vector< AbstractCardiacCellInterface* > AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mPurkinjeCellsDistributed [protected] |
The vector of the purkinje cells. Distributed. Empty unless a AbstractPurkinjeCellFactory is given to the constructor.
Definition at line 245 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue(), and AbstractCardiacTissue< SPACE_DIM >::LoadCardiacCells().
ReplicatableVector AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mPurkinjeIionicCacheReplicated [protected] |
Cache containing all the ionic currents for each purkinje node, replicated over all processes.
Definition at line 257 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue(), and AbstractCardiacTissue< SPACE_DIM >::load().
ReplicatableVector AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::mPurkinjeIntracellularStimulusCacheReplicated [protected] |
Cache containing all the stimulus currents for each Purkinje node, replicated over all processes.
Definition at line 269 of file AbstractCardiacTissue.hpp.
Referenced by AbstractCardiacTissue< ELEMENT_DIM, SPACE_DIM >::AbstractCardiacTissue().
The documentation for this class was generated from the following files:
- heart/src/tissue/AbstractCardiacTissue.hpp
- heart/src/tissue/AbstractCardiacTissue.cpp