AbstractGeneralizedRushLarsenCardiacCell Class Reference
#include <AbstractGeneralizedRushLarsenCardiacCell.hpp>
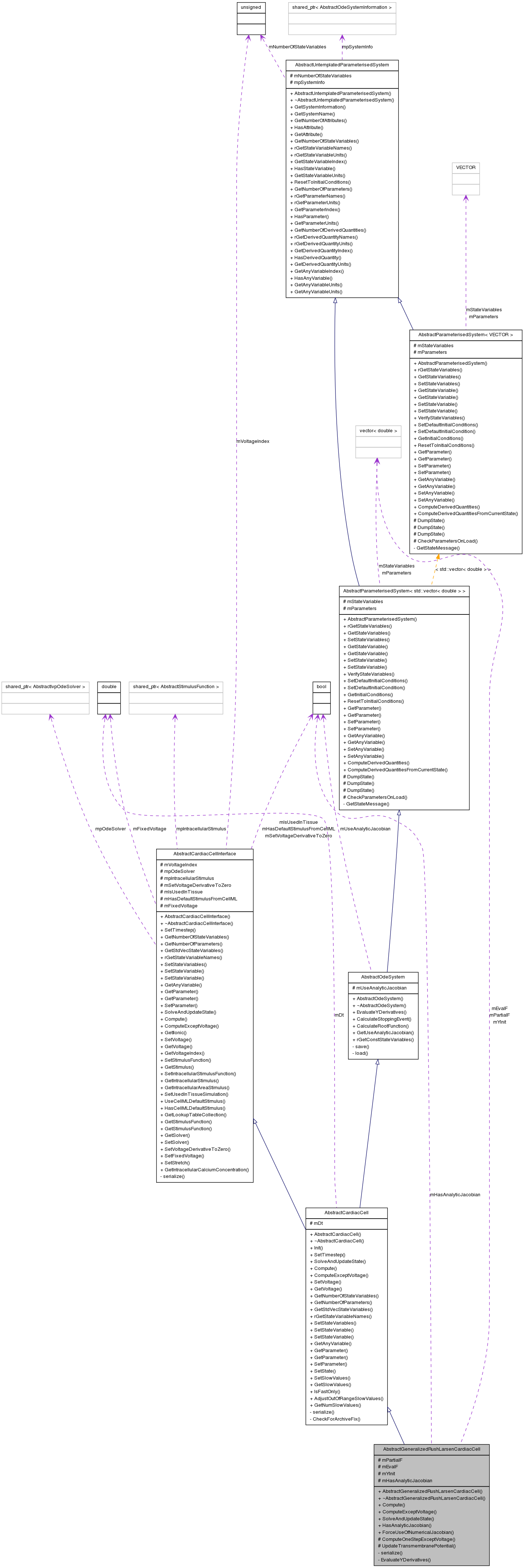
Inherits AbstractCardiacCell.
Public Member Functions | |
| AbstractGeneralizedRushLarsenCardiacCell (unsigned numberOfStateVariables, unsigned voltageIndex, boost::shared_ptr< AbstractStimulusFunction > pIntracellularStimulus) | |
| virtual | ~AbstractGeneralizedRushLarsenCardiacCell () |
| OdeSolution | Compute (double tStart, double tEnd, double tSamp=0.0) |
| void | ComputeExceptVoltage (double tStart, double tEnd) |
| void | SolveAndUpdateState (double tStart, double tEnd) |
| bool | HasAnalyticJacobian () const |
| void | ForceUseOfNumericalJacobian (bool useNumericalJacobian=true) |
Protected Member Functions | |
| virtual void | ComputeOneStepExceptVoltage (double time)=0 |
| virtual void | UpdateTransmembranePotential (double time)=0 |
Protected Attributes | |
| std::vector< double > | mPartialF |
| std::vector< double > | mEvalF |
| std::vector< double > | mYInit |
| bool | mHasAnalyticJacobian |
Private Member Functions | |
| template<class Archive > | |
| void | serialize (Archive &archive, const unsigned int version) |
| void | EvaluateYDerivatives (double time, const std::vector< double > &rY, std::vector< double > &rDY) |
Friends | |
| class | boost::serialization::access |
Detailed Description
This is the base class for cardiac cells solved using the GRL methods (GRL1 and GRL2). Modified from AbstractRushLarsenCardiacCell.hpp
Definition at line 59 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
Constructor & Destructor Documentation
| AbstractGeneralizedRushLarsenCardiacCell::AbstractGeneralizedRushLarsenCardiacCell | ( | unsigned | numberOfStateVariables, | |
| unsigned | voltageIndex, | |||
| boost::shared_ptr< AbstractStimulusFunction > | pIntracellularStimulus | |||
| ) |
Standard constructor for a cell.
- Parameters:
-
numberOfStateVariables the size of the ODE system voltageIndex the index of the variable representing the transmembrane potential within the state variable vector pIntracellularStimulus the intracellular stimulus function
Some notes for future reference:
- It's a pity that inheriting from AbstractCardiacCell forces us to store a null pointer (for the unused ODE solver) in every instance. We may want to revisit this design decision at a later date.
Definition at line 53 of file AbstractGeneralizedRushLarsenCardiacCell.cpp.
| AbstractGeneralizedRushLarsenCardiacCell::~AbstractGeneralizedRushLarsenCardiacCell | ( | ) | [virtual] |
Virtual destructor
Definition at line 67 of file AbstractGeneralizedRushLarsenCardiacCell.cpp.
Member Function Documentation
| OdeSolution AbstractGeneralizedRushLarsenCardiacCell::Compute | ( | double | tStart, | |
| double | tEnd, | |||
| double | tSamp = 0.0 | |||
| ) | [virtual] |
Simulates this cell's behaviour between the time interval [tStart, tEnd], with timestep mDt.
The length of the time interval must be a multiple of the timestep.
- Parameters:
-
tStart beginning of the time interval to simulate tEnd end of the time interval to simulate tSamp sampling interval for returned results (defaults to mDt)
- Returns:
- the values of each state variable, at intervals of tSamp.
Reimplemented from AbstractCardiacCell.
Definition at line 70 of file AbstractGeneralizedRushLarsenCardiacCell.cpp.
References ComputeOneStepExceptVoltage(), AbstractCardiacCellInterface::GetVoltageIndex(), AbstractCardiacCell::mDt, AbstractUntemplatedParameterisedSystem::mpSystemInfo, OdeSolution::rGetSolutions(), AbstractParameterisedSystem< std::vector< double > >::rGetStateVariables(), OdeSolution::rGetTimes(), OdeSolution::SetNumberOfTimeSteps(), OdeSolution::SetOdeSystemInformation(), UpdateTransmembranePotential(), and AbstractParameterisedSystem< std::vector< double > >::VerifyStateVariables().
| void AbstractGeneralizedRushLarsenCardiacCell::ComputeExceptVoltage | ( | double | tStart, | |
| double | tEnd | |||
| ) | [virtual] |
Simulates this cell's behaviour between the time interval [tStart, tEnd], with timestep mDt. The transmembrane potential is kept fixed throughout, but the other state variables are updated.
The length of the time interval must be a multiple of the timestep.
- Parameters:
-
tStart beginning of the time interval to simulate tEnd end of the time interval to simulate
Reimplemented from AbstractCardiacCell.
Definition at line 115 of file AbstractGeneralizedRushLarsenCardiacCell.cpp.
References TimeStepper::AdvanceOneTimeStep(), ComputeOneStepExceptVoltage(), TimeStepper::GetTime(), TimeStepper::IsTimeAtEnd(), AbstractCardiacCell::mDt, AbstractCardiacCellInterface::SetVoltageDerivativeToZero(), and AbstractParameterisedSystem< std::vector< double > >::VerifyStateVariables().
| virtual void AbstractGeneralizedRushLarsenCardiacCell::ComputeOneStepExceptVoltage | ( | double | time | ) | [protected, pure virtual] |
Update the values of all variables except the transmembrane potential using a GRL method.
- Parameters:
-
time the current simulation time
Referenced by Compute(), ComputeExceptVoltage(), and SolveAndUpdateState().
| void AbstractGeneralizedRushLarsenCardiacCell::EvaluateYDerivatives | ( | double | time, | |
| const std::vector< double > & | rY, | |||
| std::vector< double > & | rDY | |||
| ) | [inline, private, virtual] |
This function should never be called - the cell class incorporates its own solver.
- Parameters:
-
time rY rDY
Implements AbstractOdeSystem.
Definition at line 157 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
References NEVER_REACHED.
| void AbstractGeneralizedRushLarsenCardiacCell::ForceUseOfNumericalJacobian | ( | bool | useNumericalJacobian = true |
) |
Force the use of a numerical Jacobian, even if an analytic form is provided. This is needed for a handful of troublesome models.
- Parameters:
-
useNumericalJacobian Whether to use a numerical instead of the analytic Jacobian.
Definition at line 159 of file AbstractGeneralizedRushLarsenCardiacCell.cpp.
References EXCEPTION, and AbstractOdeSystem::mUseAnalyticJacobian.
| bool AbstractGeneralizedRushLarsenCardiacCell::HasAnalyticJacobian | ( | ) | const |
- Returns:
- whether the ODE system has an analytic Jacobian (mHasAnalyticJacobian).
Definition at line 154 of file AbstractGeneralizedRushLarsenCardiacCell.cpp.
References mHasAnalyticJacobian.
| void AbstractGeneralizedRushLarsenCardiacCell::serialize | ( | Archive & | archive, | |
| const unsigned int | version | |||
| ) | [inline, private] |
Archive the member variables.
- Parameters:
-
archive version
Reimplemented from AbstractCardiacCell.
Definition at line 71 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
| void AbstractGeneralizedRushLarsenCardiacCell::SolveAndUpdateState | ( | double | tStart, | |
| double | tEnd | |||
| ) | [virtual] |
Simulate this cell's behaviour between the time interval [tStart, tEnd], with timestemp mDt, updating the internal state variable values.
- Parameters:
-
tStart beginning of the time interval to simulate tEnd end of the time interval to simulate
Reimplemented from AbstractCardiacCell.
Definition at line 134 of file AbstractGeneralizedRushLarsenCardiacCell.cpp.
References TimeStepper::AdvanceOneTimeStep(), ComputeOneStepExceptVoltage(), TimeStepper::GetTime(), AbstractCardiacCellInterface::GetVoltageIndex(), TimeStepper::IsTimeAtEnd(), AbstractCardiacCell::mDt, AbstractParameterisedSystem< std::vector< double > >::rGetStateVariables(), UpdateTransmembranePotential(), and AbstractParameterisedSystem< std::vector< double > >::VerifyStateVariables().
| virtual void AbstractGeneralizedRushLarsenCardiacCell::UpdateTransmembranePotential | ( | double | time | ) | [protected, pure virtual] |
Perform a forward Euler step to update the transmembrane potential.
- Parameters:
-
time the current simulation time
Referenced by Compute(), and SolveAndUpdateState().
Friends And Related Function Documentation
friend class boost::serialization::access [friend] |
Needed for serialization.
Reimplemented from AbstractCardiacCell.
Definition at line 63 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
Member Data Documentation
std::vector<double> AbstractGeneralizedRushLarsenCardiacCell::mEvalF [protected] |
The derivatives, working memory for use by subclasses.
Definition at line 182 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
Referenced by AbstractGeneralizedRushLarsenCardiacCell().
Whether we have an analytic Jacobian.
Definition at line 188 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
Referenced by HasAnalyticJacobian().
std::vector<double> AbstractGeneralizedRushLarsenCardiacCell::mPartialF [protected] |
The diagonal of the Jacobian, working memory for use by subclasses.
Definition at line 179 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
Referenced by AbstractGeneralizedRushLarsenCardiacCell().
std::vector<double> AbstractGeneralizedRushLarsenCardiacCell::mYInit [protected] |
The state at the beginning of the current step, working memory for use by subclasses.
Definition at line 185 of file AbstractGeneralizedRushLarsenCardiacCell.hpp.
Referenced by AbstractGeneralizedRushLarsenCardiacCell().
The documentation for this class was generated from the following files:
- heart/src/odes/AbstractGeneralizedRushLarsenCardiacCell.hpp
- heart/src/odes/AbstractGeneralizedRushLarsenCardiacCell.cpp
 1.6.2
1.6.2